
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,982,444 – 20,982,647 |
| Length | 203 |
| Max. P | 0.990474 |

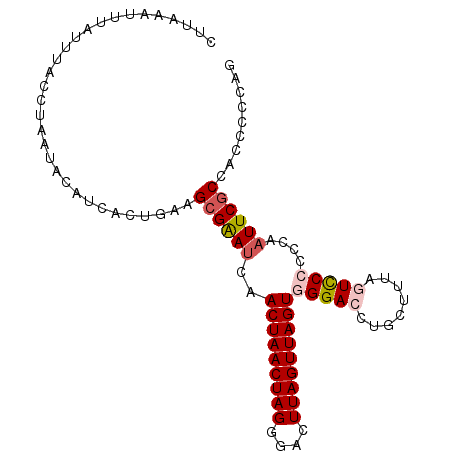
| Location | 20,982,444 – 20,982,542 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982444 98 + 23771897 CUUAAAUUUAUUUAACUAAUACAUCACUGAAGCGAAUCAACUAACUAGGGACUUAGUUAGUGGGACCAGCUUUAGUCCCCCCAAUUCGCCACCCCCAG ...............................((((((..(((((((((....)))))))))(((((........)))))....))))))......... ( -22.60) >DroSec_CAF1 113 98 + 1 CUUAAAUUUAUUUACCUAAUACAUCACUGAAGCGAAUCGACUAACUAGGGACUUAGUUAGUGGGACCUGCUUUACUCCACCCAAUUCGCCACCCCCAG ...............................((((((..(((((((((....)))))))))(((...............))).))))))......... ( -17.76) >DroSim_CAF1 7511 98 + 1 CUUAAAUUUAUUUACCUAAUACAUCACUGAAGCGAAUCGACUAACUAGGGACUUAGUUAGUGGGACCUGCUUUAGUCCACCCAAUUCGCCACCCCCAG .........................(((((((((..((.(((((((((....))))))))).))...)))))))))...................... ( -21.30) >DroEre_CAF1 3207 98 + 1 CUUAAAUUUAUUUACCUAACACCUAAUUGAAGCGGAUCAACUAACUAGGGACUUAGUUAGUGGGACCUCUUUUAGUCCCCUCAAUUCGCCACCCCCAC ...............................((((((..(((((((((....)))))))))(((((........)))))....))))))......... ( -22.10) >DroYak_CAF1 6204 98 + 1 CUUAAAUUUAUUUACCUAAUACAUAAUUGAAGAGGAGCAACUAACUAGGGAAUUAGUUAGUGGGAGCUGCUUAAGUUCCCCCAAUUCGCCACCCCCAG ............................(((..((....(((((((((....)))))))))(((((((.....)))))))))..)))........... ( -23.60) >consensus CUUAAAUUUAUUUACCUAAUACAUCACUGAAGCGAAUCAACUAACUAGGGACUUAGUUAGUGGGACCUGCUUUAGUCCCCCCAAUUCGCCACCCCCAG ...............................((((((..(((((((((....)))))))))(((((........)))))....))))))......... (-16.44 = -17.24 + 0.80)


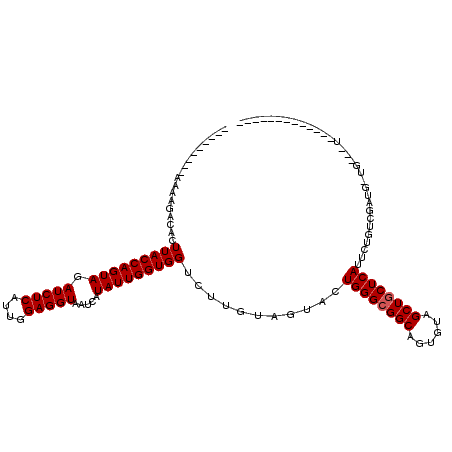
| Location | 20,982,542 – 20,982,647 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982542 105 - 23771897 ---------AAAAGCCACUUACCAGUAAAUCUCGUUGGAGGUAAUCAUAUUGGUGGUCUUGUAAUACUGGGCGGCAGUGUAGCUGCUCAUUCUGCCG-UG-UGUUUUUGUUGUUUU---- ---------((((((((((((((((((.(((((....))))).....)))))))))...........((((((((......)))))))).......)-))-.))))))........---- ( -29.60) >DroPse_CAF1 204 106 - 1 ---------C-AGGACACUUACCAGUAGAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAGUACUGGGCGGCAGUGUAGCUGCUCAUUGUGUUUCAGCUG---CUGUCAUUUA-CUU ---------(-(((((...((((((((.(((((....))))).....)))))))))))))).((((...(((((((((..(((.((.....)))))...))))---)))))...))-)). ( -37.30) >DroGri_CAF1 1 103 - 1 AUAUAUGUAAAGUGAGACUUACCAGUAGAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAGUAUUGGGCGGCCGUGUAGCUACUCAUUUUGUAGAUA-UA---U------------- ((((((.((..(..(((((.(((((((.(((((....))))).....))))))))))))..).....((((..((......))..)))).....)).)))-))---)------------- ( -29.50) >DroEre_CAF1 3305 107 - 1 ---------AAAAGCAACUUACCAGUAAAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAAUACUGGGCGGCAGUGUAGCUGCUCAUUCUGCCGAUG-UG---UUGUUGUUUUUUUU ---------(((((((((..........(((((....)))))...((((((((..(...........((((((((......))))))))..)..))))))-))---..)))))))))... ( -33.92) >DroAna_CAF1 1 90 - 1 ---------AG----GACUUACCAGUAUAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAGUACUGGGCGGCAGUAUAGCUGCUCAUUCUGACGGUC-UA---U------------- ---------((----((((.(((((((((((((....))))))....))))))))))))).(((...((((((((......))))))))..)))......-..---.------------- ( -31.00) >DroPer_CAF1 1 90 - 1 ---------C-AGGACACUUACCAGUAGAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAGUACUGGGCGGCAGUGUAGCUGCUCAUUGUGUUUCAG-------------------- ---------(-(((((...((((((((.(((((....))))).....))))))))))))))......((((((((......))))))))...........-------------------- ( -30.50) >consensus _________AAAAGACACUUACCAGUAGAUCUCAUUGGAGGUAAUCAUAUUGGUGGUCUUGUAGUACUGGGCGGCAGUGUAGCUGCUCAUUCUGUCGAUG_UG___U_____________ ..................(((((((((.(((((....))))).....)))))))))...........((((((((......))))))))............................... (-22.80 = -22.97 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:17 2006