
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,982,093 – 20,982,293 |
| Length | 200 |
| Max. P | 0.954564 |

| Location | 20,982,093 – 20,982,213 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982093 120 + 23771897 CGUACGUGUAUGUAUUCGACUGUCUGCCGCCUCAGCCGCAGUCGACGUCGCUGCCACCGCUUAAAUAUCCCCAAAAACGUCCCUUGAAUUCAUAUACAUAUAUAUUUUUUAUAUAUCCAU .....(((((((.((((((..(.((((.((....)).)))).)(((((............................)))))..)))))).)))))))(((((((.....))))))).... ( -26.49) >DroSim_CAF1 7164 120 + 1 CGUACGUGUAUGUAUUCGACUGUCUGCCGCCUCAGCCGCAGUCGACGUCGCUGCCGCCGCUUAAAUAUCCCCAAAAACGUCCCUUGAAUUCAUAUACAUAUAUAUUUUUUAUAUAUCCAU .....(((((((.((((((..(.((((.((....)).)))).)(((((............................)))))..)))))).)))))))(((((((.....))))))).... ( -26.49) >DroEre_CAF1 2859 120 + 1 CGUACGUGUAUGUAUUCGCCUGUCUGCCGCCUCUACCGCAGUCGACGUCGCUGCCACCGCUUAAAUAUCCCCAAAAACGUCCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUAUCCAU .....(((((((..(((....(.((((.(......).)))).)(((((............................)))))....)))..)))))))(((((((.....))))))).... ( -20.19) >DroYak_CAF1 5851 120 + 1 CGUACGUGUAUGUAUUCGCCUGUCUGCCGCCUCUACCGCAGUCGACGUCGCUGCCACCGCUUAAAAAUCCCCAAAAACGUCCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUACCCAU .....(((((((..(((....(.((((.(......).)))).)(((((............................)))))....)))..))))))).((((((.....))))))..... ( -19.49) >consensus CGUACGUGUAUGUAUUCGACUGUCUGCCGCCUCAACCGCAGUCGACGUCGCUGCCACCGCUUAAAUAUCCCCAAAAACGUCCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUAUCCAU .....(((((((.(((((...(.((((.(......).)))).)(((((............................)))))...))))).))))))).((((((.....))))))..... (-21.32 = -21.82 + 0.50)


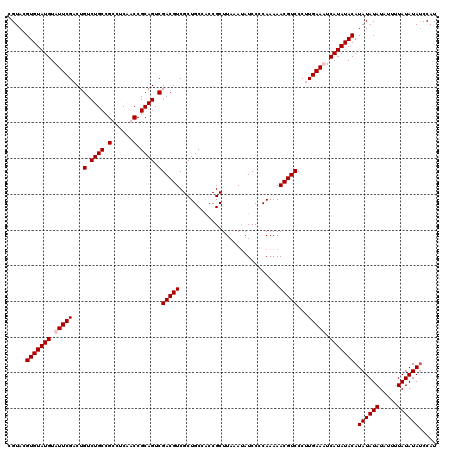
| Location | 20,982,133 – 20,982,253 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -30.61 |
| Energy contribution | -30.55 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982133 120 - 23771897 UUUCUUUUCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGGACGUUUUUGGGGAUAUUUAAGCGGUGGCAGCGACGUCGAC .......((((...((((((((((.((((.((((((....(((.(((((((.....))))))).)))....)))))).)).))...))))))))))....))))((((......)))).. ( -30.80) >DroSim_CAF1 7204 116 - 1 U----UUUCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGGACGUUUUUGGGGAUAUUUAAGCGGCGGCAGCGACGUCGAC .----..((((...((((((((((.((((.((((((....(((.(((((((.....))))))).)))....)))))).)).))...))))))))))....))))((((......)))).. ( -32.50) >DroEre_CAF1 2899 116 - 1 -U---UUCCGCGUUAUAUUUUUAAUCGCUGUUGAGUGUAGAUGGAUAUAUAAAAUAUAUAUAUGUAUAUGAUUUCAAGGGACGUUUUUGGGGAUAUUUAAGCGGUGGCAGCGACGUCGAC -.---..((((...((((((((((.((((.(((((.....(((.(((((((.....))))))).))).....))))).)).))...))))))))))....))))((((......)))).. ( -29.20) >DroYak_CAF1 5891 117 - 1 UU---UUCCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGGUAUAUAAAAUAUAUAUAUGUAUAUGAUUUCAAGGGACGUUUUUGGGGAUUUUUAAGCGGUGGCAGCGACGUCGAC ..---..((((.....((((((((.((((.(((((.....(((.(((((((.....))))))).))).....))))).)).))...))))))))......))))((((......)))).. ( -28.50) >consensus UU___UUCCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGGACGUUUUUGGGGAUAUUUAAGCGGUGGCAGCGACGUCGAC .......((((...((((((((((.((((.((((((....(((.(((((((.....))))))).)))....)))))).)).))...))))))))))....))))((((......)))).. (-30.61 = -30.55 + -0.06)


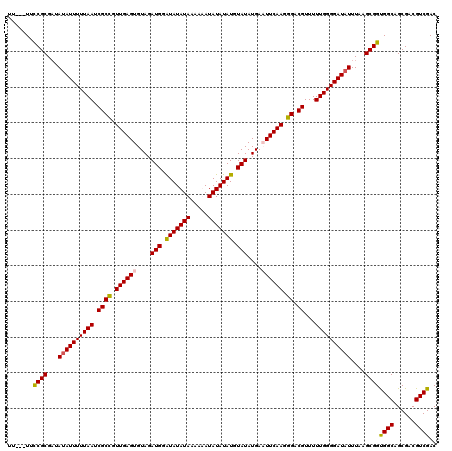
| Location | 20,982,173 – 20,982,293 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -14.43 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982173 120 + 23771897 CCCUUGAAUUCAUAUACAUAUAUAUUUUUUAUAUAUCCAUCUACACUCAACGGCGAUUAAAAAUAUAUCGCGAAAAGAAAAAAUAUAAAAACGAACUUCAAAGUGAAAUUCGAAAUUCCU .....(((((..........((((((((((...............(.....)(((((.........)))))......))))))))))....((((.(((.....))).)))).))))).. ( -15.40) >DroSim_CAF1 7244 116 + 1 CCCUUGAAUUCAUAUACAUAUAUAUUUUUUAUAUAUCCAUCUACACUCAACGGCGAUUAAAAAUAUAUCGCGAAA----AAAACAUAAAAACAAACUUCAAAGUGAAAUUCGAAAUUCCU ...((((((((((....(((((((.....)))))))................(((((.........)))))....----.......................))).)))))))....... ( -14.40) >DroEre_CAF1 2939 116 + 1 CCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUAUCCAUCUACACUCAACAGCGAUUAAAAAUAUAACGCGGAA---A-AAAUAUAAAAACGAACUUCAAAGUGAAAUUCGAAAUUCCU .((((((..........(((((((.....)))))))..........))))..(((.(((......))))))))..---.-...........((((.(((.....))).))))........ ( -13.45) >DroYak_CAF1 5931 117 + 1 CCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUACCCAUCUACACUCAACGGCGAUUAAAAAUAUAUCGCGGAA---AAAAAUAUAAAAACGAACUUCAAAGUGAAAUUCGAAAUUCCU .((((((...........((((((.....))))))...........))))..(((((.........)))))))..---.............((((.(((.....))).))))........ ( -14.45) >consensus CCCUUGAAAUCAUAUACAUAUAUAUAUUUUAUAUAUCCAUCUACACUCAACGGCGAUUAAAAAUAUAUCGCGAAA___AAAAAUAUAAAAACGAACUUCAAAGUGAAAUUCGAAAUUCCU ...((((((((((.....((((((.....)))))).................(((((.........)))))...............................))).)))))))....... (-11.20 = -11.95 + 0.75)


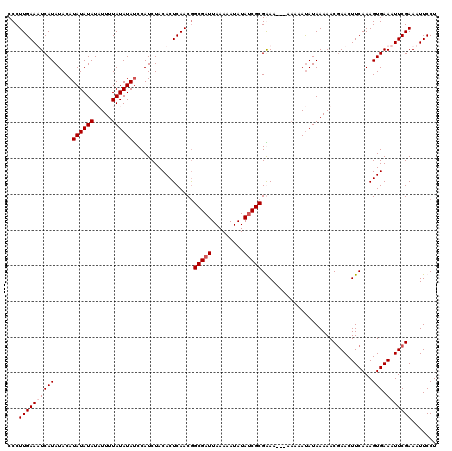
| Location | 20,982,173 – 20,982,293 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -23.75 |
| Energy contribution | -23.69 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20982173 120 - 23771897 AGGAAUUUCGAAUUUCACUUUGAAGUUCGUUUUUAUAUUUUUUCUUUUCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGG ..((((((((((......)))))))))).....................(((((.........)))))(.((((((....(((.(((((((.....))))))).)))....)))))).). ( -26.20) >DroSim_CAF1 7244 116 - 1 AGGAAUUUCGAAUUUCACUUUGAAGUUUGUUUUUAUGUUUU----UUUCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGG ..((((((((((......)))))))))).............----....(((((.........)))))(.((((((....(((.(((((((.....))))))).)))....)))))).). ( -24.00) >DroEre_CAF1 2939 116 - 1 AGGAAUUUCGAAUUUCACUUUGAAGUUCGUUUUUAUAUUU-U---UUCCGCGUUAUAUUUUUAAUCGCUGUUGAGUGUAGAUGGAUAUAUAAAAUAUAUAUAUGUAUAUGAUUUCAAGGG .(((((.....))))).(((((((((.(((..((((((((-.---..(.((((((......))).))).)..))))))))(((.(((((((.....))))))).))))))))))))))). ( -24.00) >DroYak_CAF1 5931 117 - 1 AGGAAUUUCGAAUUUCACUUUGAAGUUCGUUUUUAUAUUUUU---UUCCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGGUAUAUAAAAUAUAUAUAUGUAUAUGAUUUCAAGGG .((((...((((((((.....)))))))).............---))))(((((.........)))))(.(((((.....(((.(((((((.....))))))).))).....))))).). ( -26.09) >consensus AGGAAUUUCGAAUUUCACUUUGAAGUUCGUUUUUAUAUUUUU___UUCCGCGAUAUAUUUUUAAUCGCCGUUGAGUGUAGAUGGAUAUAUAAAAAAUAUAUAUGUAUAUGAAUUCAAGGG .((((...((((((((.....))))))))................))))(((((.........)))))(.((((((....(((.(((((((.....))))))).)))....)))))).). (-23.75 = -23.69 + -0.06)

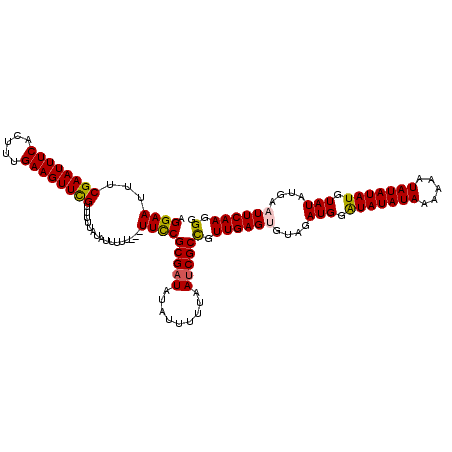
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:15 2006