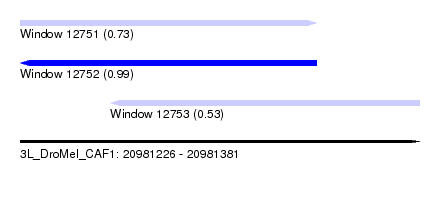
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,981,226 – 20,981,381 |
| Length | 155 |
| Max. P | 0.988874 |

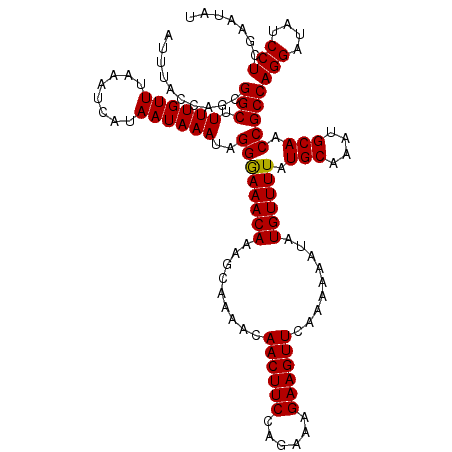
| Location | 20,981,226 – 20,981,341 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20981226 115 + 23771897 AUUCGCAUUUUGCAAGGAUAUUCGAGAAGGGGAGAAUAUUCGAGGAUAUCCUGGCGGUUGCAUUUGCAUAAAACAUAUUUUUUGAACUUCUUUCUGGAAGUUGUUUUGCUUUGUU ....(((..((((.(((((((((..(((..........)))..))))))))).)))).)))....(((..(((((.(((((..(((.....)))..)))))))))))))...... ( -29.70) >DroSim_CAF1 6314 113 + 1 AUUCGCAUUUUGCAAGGAUAUUCGAGAAGGGG--AAUAUUCGAGGAUAUCCUGGCGGUUGCAUUUGCAUAAAACAUAUUUUCUGAACUUCUUUCUGGAAGUUGUUUUGCUUUGUU ....(((..((((.(((((((((..(((....--....)))..))))))))).)))).)))....(((..(((((.((((((.(((.....))).))))))))))))))...... ( -31.90) >DroEre_CAF1 2010 113 + 1 AUUCGCAUUUUGCAAGGAUAUUCAAGAAGGGG--AAUAUUUGAGGACAUCCUGGCGGUUGCAUUUGCAUAAAACAUAUUUUUCGAACUUCUUUCUGGAAGUUGUUUUGCUUUGUU ....(((...(((((((((((((........)--))))))).(((....))).....)))))..))).(((((((..........((((((....)))))))))))))....... ( -24.30) >consensus AUUCGCAUUUUGCAAGGAUAUUCGAGAAGGGG__AAUAUUCGAGGAUAUCCUGGCGGUUGCAUUUGCAUAAAACAUAUUUUUUGAACUUCUUUCUGGAAGUUGUUUUGCUUUGUU ....(((..((((.(((((((((..(((..........)))..))))))))).)))).)))....(((..(((((.((((((.(((.....))).))))))))))))))...... (-28.24 = -28.13 + -0.11)

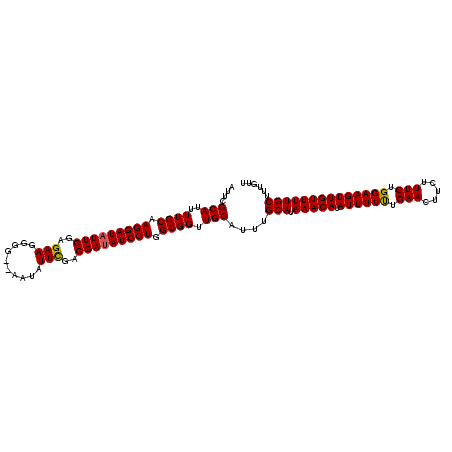

| Location | 20,981,226 – 20,981,341 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.34 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20981226 115 - 23771897 AACAAAGCAAAACAACUUCCAGAAAGAAGUUCAAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAUUCUCCCCUUCUCGAAUAUCCUUGCAAAAUGCGAAU ......(((....((((((......))))))................(((....)))...((.(((((((..((((..............))))))))))).))....))).... ( -21.94) >DroSim_CAF1 6314 113 - 1 AACAAAGCAAAACAACUUCCAGAAAGAAGUUCAGAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAUU--CCCCUUCUCGAAUAUCCUUGCAAAAUGCGAAU ......(((....((((((......))))))................(((....)))...((.(((((((..((((.....--.......))))))))))).))....))).... ( -22.10) >DroEre_CAF1 2010 113 - 1 AACAAAGCAAAACAACUUCCAGAAAGAAGUUCGAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUGUCCUCAAAUAUU--CCCCUUCUUGAAUAUCCUUGCAAAAUGCGAAU ......(((....((((((......))))))................(((....)))...((.(((((((..((((.....--.......))))))))))).))....))).... ( -20.30) >consensus AACAAAGCAAAACAACUUCCAGAAAGAAGUUCAAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAUU__CCCCUUCUCGAAUAUCCUUGCAAAAUGCGAAU ......(((....((((((......))))))................(((....)))...((.(((((((..((((..............))))))))))).))....))).... (-22.00 = -21.34 + -0.66)

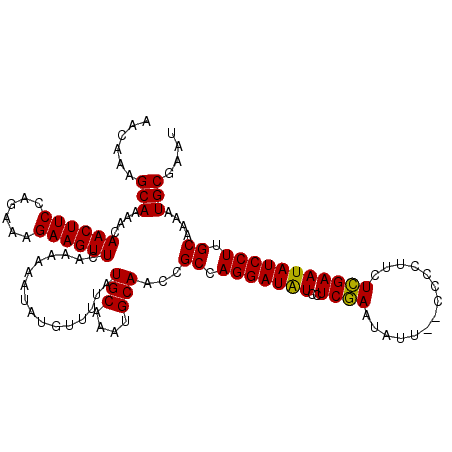

| Location | 20,981,261 – 20,981,381 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.14 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20981261 120 - 23771897 AUUUACCAGCGGCUUUUGUUUAAAUCAUAAUAAAUAGGGAAACAAAGCAAAACAACUUCCAGAAAGAAGUUCAAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAU .....((.((((..(((((.((((.((((........(....)..........((((((......)))))).......)))).)))).))))).....))))..)).............. ( -21.00) >DroSim_CAF1 6347 120 - 1 AUUUACCAGCGGCUUUUGUUUAAAUCAUAAUAAAUAGGGAAACAAAGCAAAACAACUUCCAGAAAGAAGUUCAGAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAU .....((.((((..(((((.((((.((((........(....)..........((((((......)))))).......)))).)))).))))).....))))..)).............. ( -21.00) >DroEre_CAF1 2043 120 - 1 AUUUACAAGCGGCUUUUGUUUAAAUCGUAAUAAAAAGGAAAACAAAGCAAAACAACUUCCAGAAAGAAGUUCGAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUGUCCUCAAAUAU ..........((((((((((........))))))).((((((((.........((((((......)))))).........)))))).(((....))).)))))(((....)))....... ( -20.67) >consensus AUUUACCAGCGGCUUUUGUUUAAAUCAUAAUAAAUAGGGAAACAAAGCAAAACAACUUCCAGAAAGAAGUUCAAAAAAUAUGUUUUAUGCAAAUGCAACCGCCAGGAUAUCCUCGAAUAU ..........(((.((((((........))))))..((((((((.........((((((......)))))).........)))))).(((....))).)))))(((....)))....... (-20.36 = -20.14 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:11 2006