
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,980,340 – 20,980,500 |
| Length | 160 |
| Max. P | 0.714323 |

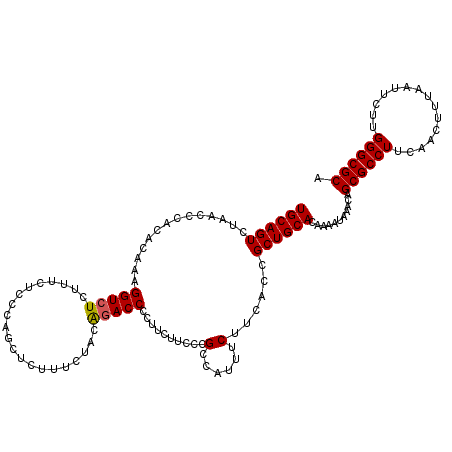
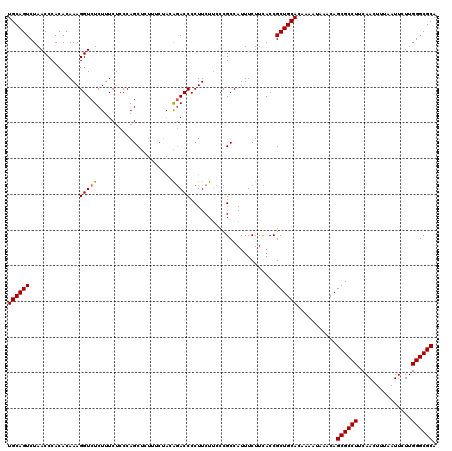
| Location | 20,980,340 – 20,980,460 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20980340 120 - 23771897 UGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACAGACCCCUUCUUCCCGCCAUUUCUUCACCGCUGCACAAAAUAAACAGCGCCUUCAACUUUAAUUCUUGGGCGCA ((((((..............(((((.....................)))))..........(......)......))))))...........((((((...............)))))). ( -18.26) >DroSim_CAF1 5449 110 - 1 UGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGC---------GGACCCCUUCU-CCCGCCAUUUCUUCACCGCUGCACAAAAUAAACAGCGCCUUCAACUUUAAUUCUUGGGCGCA ((((((...(((........)))............((---------((.........-.))))............))))))...........((((((...............)))))). ( -23.76) >DroEre_CAF1 1116 119 - 1 UGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACA-ACCCCUUCUUCCCGCCAUUUCUUUACCGCUGCACAAAAUAAACAGCGCCUUCAACUUUAAUUCUUGGGCGCA ((((((...(((........)))............((..........-.............))............))))))...........((((((...............)))))). ( -17.06) >consensus UGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACAGACCCCUUCUUCCCGCCAUUUCUUCACCGCUGCACAAAAUAAACAGCGCCUUCAACUUUAAUUCUUGGGCGCA ((((((..............(((((.....................)))))..........(......)......))))))...........((((((...............)))))). (-16.61 = -16.73 + 0.11)

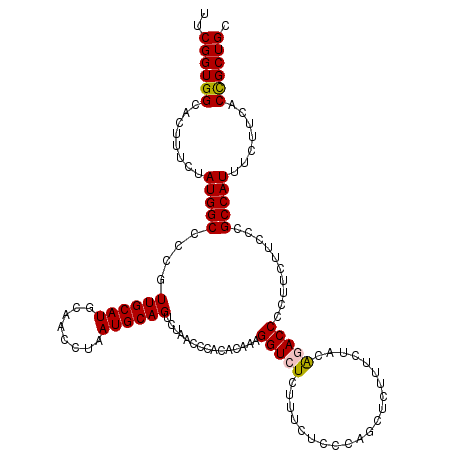
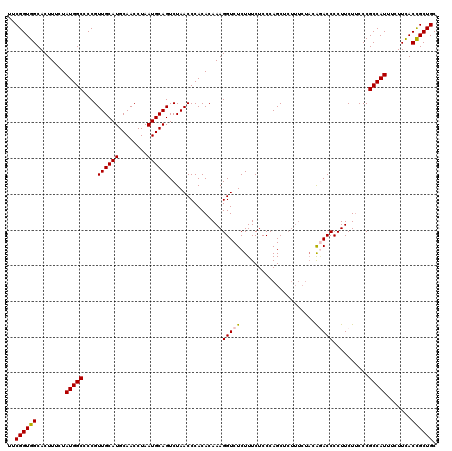
| Location | 20,980,380 – 20,980,500 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20980380 120 - 23771897 UUCGGUGGCACUUUCUAUGGCCCCGUUGCAUGCAACCUAAUGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACAGACCCCUUCUUCCCGCCAUUUCUUCACCGCUGC ..((((((........(((((....((((((........))))))...............(((((.....................)))))..........))))).......)))))). ( -23.36) >DroSim_CAF1 5489 110 - 1 AUCGGUGGCACUUUCUAUGGCCCCGUUGCAUGCAACCUAAUGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGC---------GGACCCCUUCU-CCCGCCAUUUCUUCACCGCUGC ..((((((........(((((....((((((........))))))...............(((((((.......)).---------)))))......-...))))).......)))))). ( -24.56) >DroEre_CAF1 1156 119 - 1 UUCGGUGGCACUUUCUAUGGCCCCGUUGCAUGCAACCUAAUGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACA-ACCCCUUCUUCCCGCCAUUUCUUUACCGCUGC ..((((((........(((((....((((((........))))))....(((........)))........................-.............))))).......)))))). ( -20.36) >DroYak_CAF1 4055 120 - 1 UUCGGUGGCACUUUCUAUGGCCCCGUUGCAUGCAACCUAAUGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACAAACCCCUUCUUCCCGCCAUUUCUUCACUGCUGC ....(..(((......(((((....((((((........))))))....(((........)))......................................)))))........)))..) ( -17.94) >consensus UUCGGUGGCACUUUCUAUGGCCCCGUUGCAUGCAACCUAAUGCAGUCUAACCCACACAAAGGUCUCUUUCUCCCAGCUCUUUCUACAGACCCCUUCUUCCCGCCAUUUCUUCACCGCUGC ..((((((........(((((....((((((........))))))...............(((((.....................)))))..........))))).......)))))). (-20.24 = -20.36 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:09 2006