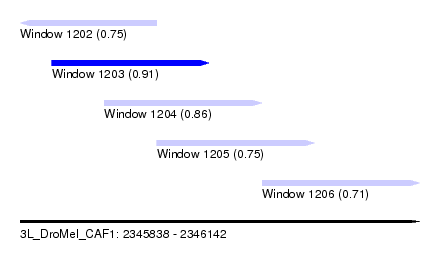
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,345,838 – 2,346,142 |
| Length | 304 |
| Max. P | 0.913648 |

| Location | 2,345,838 – 2,345,942 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -30.90 |
| Energy contribution | -32.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2345838 104 - 23771897 CAUCUGGACCUGGUCAUAUACGACCAUGUCAGUGCAGAGUUUCACCAUGAUGGGUGCCACCGCCGCAUUGUGAAUAAAGGCGGAGAGCAGGGCCGAAAGCCAGA (((((((((.(((((......))))).))).(((........))))).))))..((((.(((((..............))))).).))).((.(....)))... ( -32.84) >DroSec_CAF1 94930 104 - 1 UAUCUGGCCCUCGUCAUAUACGACCAUGUCAGUGCAGAGUUUCACCAUGAUGGGGGCCACCGCCGCAUUGUGGAUAAAGGCGGAGAGCAGGGCCGAAAGCCAGA ..(((((((((((((((....(((..(((....)))..))).....)))))))))))).(((((..............))))))))....((.(....)))... ( -39.14) >DroEre_CAF1 101533 104 - 1 UAUCUGCACCUCAUCAUGGAUGACCAUGUCAGUGCAGAGUUUCACCAUGAUGGGGGCCACCGCCGCAUUGUGGAUGAAGGCGGAGAGCAGGGCUGAAAGCCAGA ...((((.(((((((((((..(((..(((....)))..)))...)))))))))))....(((((.((((...))))..)))))...))))(((.....)))... ( -43.80) >DroYak_CAF1 98068 104 - 1 UAUCUGCACCUCAUCAUGGACGACCAUGUCGGUGCAGAGUUUCACCAUGAUGGGGGCCACCGCCGCAUUGUGGAUAAAGGCGGAGAGCAGGGACGAAAGCCAGA ...((((.(((((((((((..(((..(((....)))..)))...)))))))))))....(((((..............)))))...))))((.(....)))... ( -41.54) >consensus UAUCUGCACCUCAUCAUAGACGACCAUGUCAGUGCAGAGUUUCACCAUGAUGGGGGCCACCGCCGCAUUGUGGAUAAAGGCGGAGAGCAGGGCCGAAAGCCAGA ...((((.(((((((((((..(((..(((....)))..)))...)))))))))))....(((((..............)))))...))))((.(....)))... (-30.90 = -32.47 + 1.56)

| Location | 2,345,862 – 2,345,982 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -36.83 |
| Energy contribution | -37.45 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2345862 120 + 23771897 CCUUUAUUCACAAUGCGGCGGUGGCACCCAUCAUGGUGAAACUCUGCACUGACAUGGUCGUAUAUGACCAGGUCCAGAUGCCCCUACUGCCGCUCAUCUGGGCCACAAGCAUGGGCACCA ..............((((((((((((((......)))).......((((((((.((((((....)))))).)).))).)))..))))))))))......(((((.((....))))).)). ( -45.10) >DroSec_CAF1 94954 120 + 1 CCUUUAUCCACAAUGCGGCGGUGGCCCCCAUCAUGGUGAAACUCUGCACUGACAUGGUCGUAUAUGACGAGGGCCAGAUACGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCA ..............(((((((((((((((((...((((........))))...))))((((.....)))))))))((......))))))))))......(((((.(......)))).)). ( -40.30) >DroEre_CAF1 101557 120 + 1 CCUUCAUCCACAAUGCGGCGGUGGCCCCCAUCAUGGUGAAACUCUGCACUGACAUGGUCAUCCAUGAUGAGGUGCAGAUACGCCUACUGCCCCUCAUCUGGGCCACGAGCAUGGGCACCA .......((...((((..((.((((((((((...((((........))))...))))........(((((((.((((.........)))).))))))).)))))))).)))).))..... ( -43.70) >DroYak_CAF1 98092 120 + 1 CCUUUAUCCACAAUGCGGCGGUGGCCCCCAUCAUGGUGAAACUCUGCACCGACAUGGUCGUCCAUGAUGAGGUGCAGAUACGCCUACUGCCGCUCAUCUGGGCCACCAGCAUGGGUACCA ....((((((....(((((((((((...(((....)))....((((((((..(((((....)))))....))))))))...))).))))))))....((((....))))..))))))... ( -52.60) >consensus CCUUUAUCCACAAUGCGGCGGUGGCCCCCAUCAUGGUGAAACUCUGCACUGACAUGGUCGUACAUGACGAGGUCCAGAUACGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCA ..............(((((((((((...(((....)))....((((((((..((((......))))....))))))))...))).))))))))......(((((.(......)))).)). (-36.83 = -37.45 + 0.62)

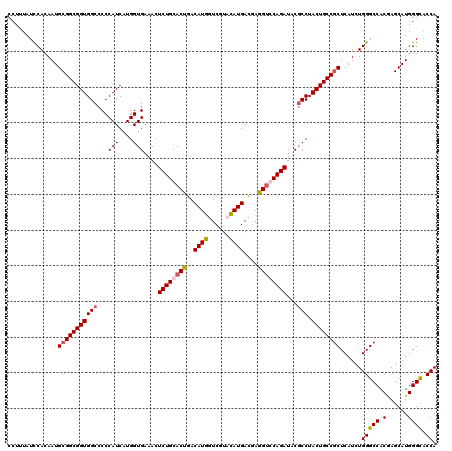
| Location | 2,345,902 – 2,346,022 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -47.27 |
| Consensus MFE | -36.67 |
| Energy contribution | -37.05 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2345902 120 + 23771897 ACUCUGCACUGACAUGGUCGUAUAUGACCAGGUCCAGAUGCCCCUACUGCCGCUCAUCUGGGCCACAAGCAUGGGCACCAGCUACGGUGCCAAUGGCACUCUCUUGGGCUCCCUGGCCAA ..............((((((....))))))((.((((..((((....((((((((....))))..........((((((......))))))...)))).......))))...)))))).. ( -44.90) >DroSec_CAF1 94994 120 + 1 ACUCUGCACUGACAUGGUCGUAUAUGACGAGGGCCAGAUACGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGUGCCAAUGGCACUCUCUUGGGCUCCCUGGCCAA .(((....(......)((((....)))))))((((((....(((((.((((((((...........))))...((((((......))))))...))))......)))))...)))))).. ( -43.70) >DroEre_CAF1 101597 120 + 1 ACUCUGCACUGACAUGGUCAUCCAUGAUGAGGUGCAGAUACGCCUACUGCCCCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGAGCCAAUGGCACCCUCUUGGGCUCCUUGGCCAA ..((((((((..(((((....)))))....))))))))...(((((.((((((......)))......))))))))....((((.((((((...((...)).....)))))).))))... ( -48.90) >DroYak_CAF1 98132 120 + 1 ACUCUGCACCGACAUGGUCGUCCAUGAUGAGGUGCAGAUACGCCUACUGCCGCUCAUCUGGGCCACCAGCAUGGGUACCAGCUAUGGAGCCAAUGGCACCCUCUUGGGCUCCCUGGCCAA ..((((((((..(((((....)))))....))))))))...(((..(((..((((((((((....)))).))))))..)))....((((((...((...)).....))))))..)))... ( -51.60) >consensus ACUCUGCACUGACAUGGUCGUACAUGACGAGGUCCAGAUACGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGAGCCAAUGGCACCCUCUUGGGCUCCCUGGCCAA ..((((((((..((((......))))....))))))))......................(((((...((...((((((......))))))....)).(((....))).....))))).. (-36.67 = -37.05 + 0.38)

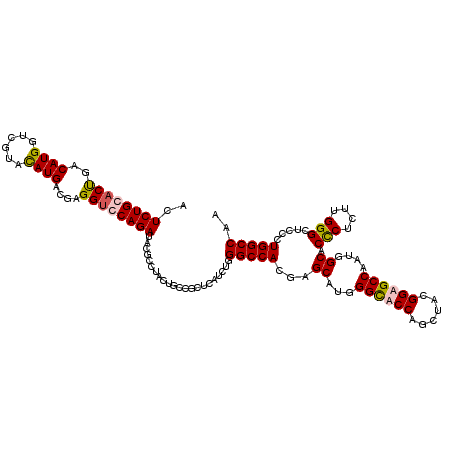
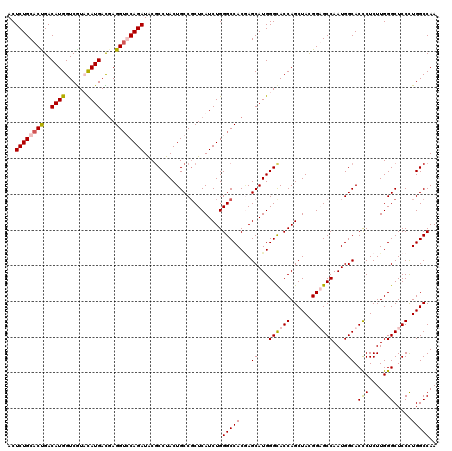
| Location | 2,345,942 – 2,346,062 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -51.33 |
| Consensus MFE | -47.33 |
| Energy contribution | -48.20 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2345942 120 + 23771897 CCCCUACUGCCGCUCAUCUGGGCCACAAGCAUGGGCACCAGCUACGGUGCCAAUGGCACUCUCUUGGGCUCCCUGGCCAACGAGUUCGCCGCGGUGGUGGGCAAAGCUCAUGGCUACAAG ..(((((..((((((....)))).....((...((((((......))))))...(((...(((((((.(.....).)))).)))...)))))))..)))))...(((.....)))..... ( -50.40) >DroSec_CAF1 95034 120 + 1 CGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGUGCCAAUGGCACUCUCUUGGGCUCCCUGGCCAACGAGUUCGUCGCGGUGGUGGGCAAAGCUCAUGGCUACAAG .((((((..((((.......(((((.((((((.((((((......)))))).)))...)))....((....))))))).(((....))).))))..))))))..(((.....)))..... ( -54.00) >DroEre_CAF1 101637 120 + 1 CGCCUACUGCCCCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGAGCCAAUGGCACCCUCUUGGGCUCCUUGGCCAACGAGUUCGUCUCGGUGGUGGGCAGAGCUUAUGGCUACAAG .(((..((((((.(((((..(((((..(((.(((...))))))..((((((...((...)).....)))))).)))))...(((.....)))))))).)))))).......)))...... ( -48.50) >DroYak_CAF1 98172 120 + 1 CGCCUACUGCCGCUCAUCUGGGCCACCAGCAUGGGUACCAGCUAUGGAGCCAAUGGCACCCUCUUGGGCUCCCUGGCCAACGAGUUCGUCGCGGUGGUGGGCAGGGCUCAUGGCUACAAG .((((((..((((.(((((((....)))).)))(((.(((....))).)))...(((...(((((((.(.....).)))).)))...)))))))..))))))..(((.....)))..... ( -52.40) >consensus CGCCUACUGCCGCUCAUCUGGGCCACGAGCAUGGGCACCAGCUACGGAGCCAAUGGCACCCUCUUGGGCUCCCUGGCCAACGAGUUCGUCGCGGUGGUGGGCAAAGCUCAUGGCUACAAG .((((((..((((.......(((((...((...((((((......))))))....)).(((....))).....))))).(((....))).))))..))))))..(((.....)))..... (-47.33 = -48.20 + 0.87)

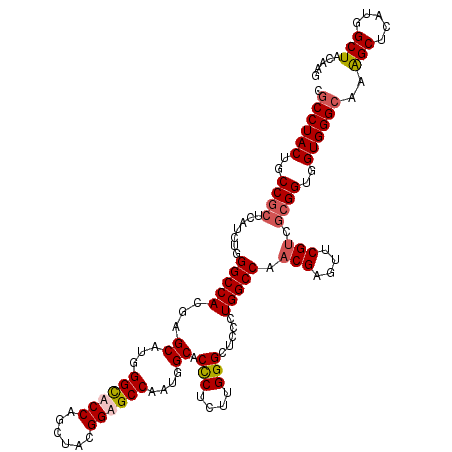
| Location | 2,346,022 – 2,346,142 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -29.19 |
| Energy contribution | -30.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2346022 120 + 23771897 CGAGUUCGCCGCGGUGGUGGGCAAAGCUCAUGGCUACAAGAUCACCUUCAGGUCAUUCUUUGUGGUUGGUUUCCUAAUCAUGCUAUUCACCGUUGUCAUCUGCUCGAUUUUCCUAAUUAU (((((..((.((((((((((((...))))))(((...((((..(((....)))...)))).((((((((....)))))))))))...)))))).)).....))))).............. ( -34.80) >DroSec_CAF1 95114 120 + 1 CGAGUUCGUCGCGGUGGUGGGCAAAGCUCAUGGCUACAAGAUCACCUUCAGGUCAUUCUUUGUGGUUGGAUUCCCUAUCAUGCUCUUCACCGUUGUCAUCUGCUCGAUUUUCCUAAUUAU (((((..(.((((((((.(((((........((((((((((..(((....)))...)).))))))))((....)).....))))).)))))).)).)....))))).............. ( -35.20) >DroEre_CAF1 101717 120 + 1 CGAGUUCGUCUCGGUGGUGGGCAGAGCUUAUGGCUACAAGAUCACCUUCCAGUCAUUCUUUGUGGUCGGUUUCCGCAUUAUGCUCUUCACCGUUGUCAUCUGCUCGAUUUUCCUAAUUAU (((((..((..(..((((((..(((((..((((((..(((.....)))..))))))....(((((.......)))))....)))))))))))..)..))..))))).............. ( -34.20) >DroYak_CAF1 98252 120 + 1 CGAGUUCGUCGCGGUGGUGGGCAGGGCUCAUGGCUACAAGAUCACUUUCAGGGGAUUCUUUGUGGUAGGAUUCCGCAUUAUGCUCUUCACCGUUGUCGUCUGCUCGAUAUUCUUAAUUAU (((((.((.((((((((.(((((((..((...(((((((((((.((....)).))))..)))))))..))..))......))))).)))))).)).))...))))).............. ( -39.60) >consensus CGAGUUCGUCGCGGUGGUGGGCAAAGCUCAUGGCUACAAGAUCACCUUCAGGUCAUUCUUUGUGGUUGGAUUCCGCAUCAUGCUCUUCACCGUUGUCAUCUGCUCGAUUUUCCUAAUUAU (((((.....(((((((.(((((........((((((((((..(((....)))...)).))))))))((...))......))))).)))))))........))))).............. (-29.19 = -30.20 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:06 2006