| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,963,315 – 20,963,435 |
| Length | 120 |
| Max. P | 0.616837 |

| Location | 20,963,315 – 20,963,435 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -51.77 |
| Consensus MFE | -41.32 |
| Energy contribution | -40.71 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
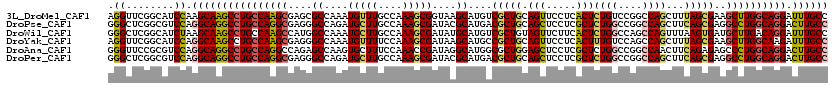
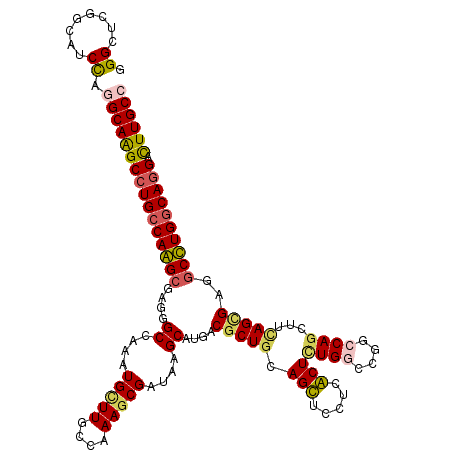
>3L_DroMel_CAF1 20963315 120 + 23771897 GGCAAAUCCUGCCAAGCUUCGCUAAAGCUGGCCGGACAGAGUGAGGAACUGCAGCGACAUGCUUACCGCUUUGGCAAACAUUUGGCGCUCGCUUGGCAGGCUUGCUUGGAUGCCGAACCU ((((...((((((((((..(((((((....((((....(((((.(.....(((......)))...)))))))))).....)))))))...))))))))))((.....)).))))...... ( -45.30) >DroPse_CAF1 220162 120 + 1 GGCAAGUCCUGCCAGGCCUCGCUGAAGCUGGCCGGCCAGAGCGAGGAGCUGCAGCGUCAUGCGUAUCGCUUUGGCAAGCAUCUGGCCCUCGCCUGGCAGGCCUGCCUGGACGCCGAGCCC ((((.(.((((((((((...(((((.(((.....((((((((((...((.(((......))))).)))))))))).))).)).)))....))))))))))).)))).((.(.....))). ( -58.90) >DroWil_CAF1 273220 120 + 1 GGCAAAUCCUGUCAAGCAUCACUUAAACUGGCUGGCCAGAGUGAAGAACUACAGCGACAUGCAUAUCGCUUUGGCAAGCAUUUGGCCAUGGCUUGGCAGGCUUGCUUAGAUGCCGAGCCC ((((((((.((((((...(((((....((((....)))))))))........(((((........))))))))))).).)))).)))..(((((((((..((.....)).))))))))). ( -43.60) >DroYak_CAF1 226506 120 + 1 GGCAAAUCUUGCCAAGCUUCGCUAAAGCUGGCUGGACAAAGUGAGGAACUGCAGCGGCAUGCUUAUCGCUUUGGAAAACAUUUGGCCCUCGCUUGGCAGGCUUGCCUGGAUGCCGAACCU (((((..((((((((((...((....)).(((..((((((((((......(((......)))...)))))))).......))..)))...)))))))))).))))).............. ( -43.71) >DroAna_CAF1 207348 120 + 1 GGCAAGUCCUGCCAGGGCUCUCUGAAGUUGGCCGGCCAGAGCGAGGAGCUCCAGCGCCAUGCCUAUCGGUUUGGAAAGCACUUGGCUCUGGCCUGGCAGGCCUGCCUGGACGCGGAACCC ((((.(.(((((((.((((..(....)..))))(((((((((.(((.(((......(((.(((....))).)))..))).))).))))))))))))))))).)))).............. ( -60.20) >DroPer_CAF1 220990 120 + 1 GGCAAGUCCUGCCAGGCCUCGCUGAAGCUGGCCGGCCAGAGCGAGGAGCUGCAGCGUCAUGCGUAUCGCUUUGGCAAGCAUCUGGCCCUCGCCUGGCAGGCCUGCCUGGACGCCGAGCCC ((((.(.((((((((((...(((((.(((.....((((((((((...((.(((......))))).)))))))))).))).)).)))....))))))))))).)))).((.(.....))). ( -58.90) >consensus GGCAAAUCCUGCCAAGCCUCGCUGAAGCUGGCCGGCCAGAGCGAGGAACUGCAGCGACAUGCGUAUCGCUUUGGCAAGCAUUUGGCCCUCGCCUGGCAGGCCUGCCUGGACGCCGAACCC ((((.(.((((((((((...((....)).(((((((((((((((......(((......)))...))))))))).......))))))...)))))))))).))))).............. (-41.32 = -40.71 + -0.61)

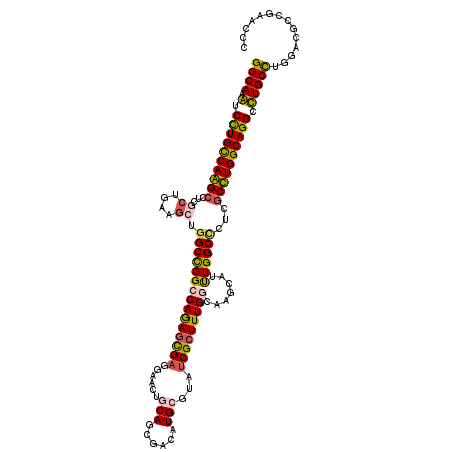
| Location | 20,963,315 – 20,963,435 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -51.67 |
| Consensus MFE | -38.62 |
| Energy contribution | -38.27 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20963315 120 - 23771897 AGGUUCGGCAUCCAAGCAAGCCUGCCAAGCGAGCGCCAAAUGUUUGCCAAAGCGGUAAGCAUGUCGCUGCAGUUCCUCACUCUGUCCGGCCAGCUUUAGCGAAGCUUGGCAGGAUUUGCC .((........))..(((((((((((((((...(((...(((((((((.....)))))))))((((..((((.........)))).))))........)))..)))))))))).))))). ( -49.00) >DroPse_CAF1 220162 120 - 1 GGGCUCGGCGUCCAGGCAGGCCUGCCAGGCGAGGGCCAGAUGCUUGCCAAAGCGAUACGCAUGACGCUGCAGCUCCUCGCUCUGGCCGGCCAGCUUCAGCGAGGCCUGGCAGGACUUGCC ((((.....)))).((((((((((((((((.(((((.((..(((.((...((((..........)))))))))..)).))))).)))((((.((....))..))))))))))).)))))) ( -63.60) >DroWil_CAF1 273220 120 - 1 GGGCUCGGCAUCUAAGCAAGCCUGCCAAGCCAUGGCCAAAUGCUUGCCAAAGCGAUAUGCAUGUCGCUGUAGUUCUUCACUCUGGCCAGCCAGUUUAAGUGAUGCUUGACAGGAUUUGCC .((((.((((.((.....))..)))).))))..(((.((((.((((.((((((((((....)))))))...((...(((((((((....))))....))))).))))).))))))))))) ( -40.00) >DroYak_CAF1 226506 120 - 1 AGGUUCGGCAUCCAGGCAAGCCUGCCAAGCGAGGGCCAAAUGUUUUCCAAAGCGAUAAGCAUGCCGCUGCAGUUCCUCACUUUGUCCAGCCAGCUUUAGCGAAGCUUGGCAAGAUUUGCC .((........)).(((((((.((((((((((((((.....))))))(((((.((..(((.(((....))))))..)).)))))........((....))...)))))))).).)))))) ( -37.40) >DroAna_CAF1 207348 120 - 1 GGGUUCCGCGUCCAGGCAGGCCUGCCAGGCCAGAGCCAAGUGCUUUCCAAACCGAUAGGCAUGGCGCUGGAGCUCCUCGCUCUGGCCGGCCAACUUCAGAGAGCCCUGGCAGGACUUGCC (((.......))).((((((((((((((((((((((...((((((((......)).))))))((.((....)).))..)))))))))((((.........).))).))))))).)))))) ( -56.40) >DroPer_CAF1 220990 120 - 1 GGGCUCGGCGUCCAGGCAGGCCUGCCAGGCGAGGGCCAGAUGCUUGCCAAAGCGAUACGCAUGACGCUGCAGCUCCUCGCUCUGGCCGGCCAGCUUCAGCGAGGCCUGGCAGGACUUGCC ((((.....)))).((((((((((((((((.(((((.((..(((.((...((((..........)))))))))..)).))))).)))((((.((....))..))))))))))).)))))) ( -63.60) >consensus GGGCUCGGCAUCCAGGCAAGCCUGCCAAGCGAGGGCCAAAUGCUUGCCAAAGCGAUAAGCAUGACGCUGCAGCUCCUCACUCUGGCCGGCCAGCUUCAGCGAGGCCUGGCAGGACUUGCC .((........)).((((((((((((((((....((....(((((....)))))....))....(((((.(((.....)))((((....))))...)))))..)))))))))).)))))) (-38.62 = -38.27 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:00 2006