| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,922,598 – 20,922,716 |
| Length | 118 |
| Max. P | 0.790394 |

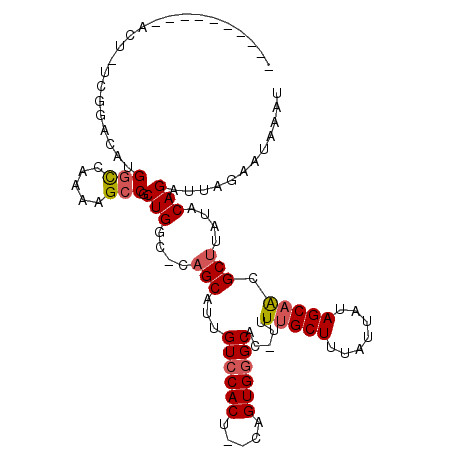
| Location | 20,922,598 – 20,922,694 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20922598 96 + 23771897 ----------ACG-UCGGACAUGGCCGAAAGCCGCUGGC-GCGCAUUGUCCACUCUCUGUGGGCCU-UUUUGCUUUAUUAUAGCAGCGCUUAUACAGAUUAGAAUAAAU ----------..(-((((....((.(....))).)))))-((((...((((((.....))))))..-...((((.......)))))))).................... ( -29.60) >DroVir_CAF1 207867 97 + 1 -------AGAACU-GCAAGCCUGGCCAAGAGCCGCUGGC-CAGCAUUGUCCACU--CAGUGGGCAC-UUUUGCUUUAUUAUAGCCAUGCUUAUACAGAUUAGAAUAAAU -------....((-(.((((.((((...(.(((...)))-)((((.(((((((.--..))))))).-...))))........)))).))))...)))............ ( -26.80) >DroGri_CAF1 189436 87 + 1 ------------------ACUUGGCCAAAAGCCGCUGGC-CAGCAUUGUCCACU--CUGUGUGCAC-UUUUGCUUUAUUAUAGCAAUGCUUAUACAGAUUAGAAUAAAU ------------------..(((((((........))))-)))..........(--(((((((((.-..(((((.......))))))))..)))))))........... ( -22.60) >DroWil_CAF1 230107 97 + 1 ----------GCUUUUGGACAUGACCAAAAGCCGCUGGCCAAGCAUUGUCCACU--CCGUGGGCCUCUUUUGCUUUAUUAUAGCAACGCUUAUACAGAUUAGAAUAAAU ----------((((((((......))))))))..(((...((((...((((((.--..)))))).....(((((.......))))).))))...)))............ ( -28.10) >DroMoj_CAF1 199172 104 + 1 AACAAGUAGAACU-GAAAAGCUGGUCAAAAGCCGCUGGC-CAGCAUUGUCCACU--CAGUGGGCAC-UUUUGCUUUAUUAUAGCAAUGCUUAUACAGAUUAGAAUAAAU ...(((((.....-.....((((((((........))))-))))..(((((((.--..))))))).-..(((((.......)))))))))).................. ( -28.30) >DroPer_CAF1 187754 94 + 1 ----------GCC-UCGGACAUGGCCAAAAGCCGCUGGC-GAGCAUUGUCCACU--CCGUGGGCUC-UUUUGCUUUAUUAUAGCAACGCUUAUACAGAUUAGAAUAAAU ----------...-(((((((.(((.....)))(((...-.)))..))))).((--..((((((..-..(((((.......))))).))))))..))....))...... ( -22.90) >consensus __________ACU_UCGGACAUGGCCAAAAGCCGCUGGC_CAGCAUUGUCCACU__CAGUGGGCAC_UUUUGCUUUAUUAUAGCAACGCUUAUACAGAUUAGAAUAAAU ......................(((.....))).(((....(((...((((((.....)))))).....(((((.......))))).)))....)))............ (-19.29 = -19.68 + 0.39)


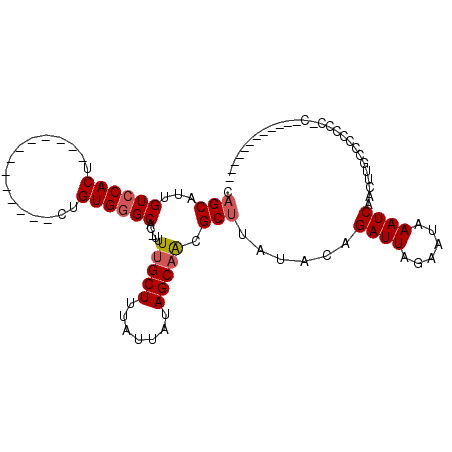
| Location | 20,922,626 – 20,922,716 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.49 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20922626 90 + 23771897 GCGCAUUGUCCACUC-----------UCUGUGGGCCU-UUUUGCUUUAUUAUAGCAGCGCUUAUACAGAUUAGAAUAAAUCAACUUGUCC-CCCGCGC-AAC----GA ((((...((((((..-----------...))))))..-..(((((.......)))))..........((((......)))).........-...))))-...----.. ( -21.40) >DroVir_CAF1 207898 81 + 1 CAGCAUUGUCCACU-------------CAGUGGGCAC-UUUUGCUUUAUUAUAGCCAUGCUUAUACAGAUUAGAAUAAAUCAACUUGCGCCCUG-C------------ .((((((((((((.-------------..))))))).-....(((.......))).)))))......((((......)))).............-.------------ ( -14.70) >DroGri_CAF1 189457 81 + 1 CAGCAUUGUCCACU-------------CUGUGUGCAC-UUUUGCUUUAUUAUAGCAAUGCUUAUACAGAUUAGAAUAAAUCAACUUGCCCCCAC-C------------ ..(((........(-------------(((((((((.-..(((((.......))))))))..)))))))...((.....))....)))......-.------------ ( -14.00) >DroWil_CAF1 230137 92 + 1 AAGCAUUGUCCACU-------------CCGUGGGCCUCUUUUGCUUUAUUAUAGCAACGCUUAUACAGAUUAGAAUAAAUCAACUUGGGCCCCAAA---AACGGUCGC ((((...((((((.-------------..)))))).....(((((.......))))).)))).....((((......))))......((((.....---...)))).. ( -20.00) >DroMoj_CAF1 199210 81 + 1 CAGCAUUGUCCACU-------------CAGUGGGCAC-UUUUGCUUUAUUAUAGCAAUGCUUAUACAGAUUAGAAUAAAUCAACUGGCCCCCUC-U------------ .((((.(((((((.-------------..))))))).-..(((((.......)))))))))......((((......)))).............-.------------ ( -16.60) >DroAna_CAF1 175801 103 + 1 GAGCAUUGUCCACCCAAUGGACCCAGUCUGUGGGCGC-UUUUGCUUUAUUAUAGCAGCGCUUAUACAGAUUAGAAUAAAUCAACUUGUCCCCCCGCUAGGAG----GA (..((..(((((.....)))))..(((((((((((((-(...(((.......)))))))))..))))))))..............))..)((((....)).)----). ( -28.60) >consensus CAGCAUUGUCCACU_____________CUGUGGGCAC_UUUUGCUUUAUUAUAGCAACGCUUAUACAGAUUAGAAUAAAUCAACUUGCCCCCCC_C____________ .(((...((((((................)))))).....(((((.......))))).)))......((((......))))........................... (-13.21 = -13.49 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:52 2006