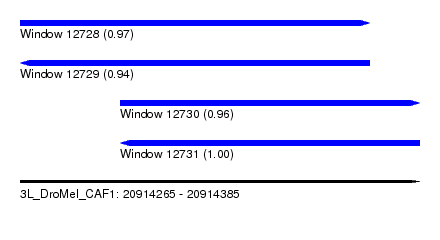
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,914,265 – 20,914,385 |
| Length | 120 |
| Max. P | 0.995678 |

| Location | 20,914,265 – 20,914,370 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -10.08 |
| Energy contribution | -11.16 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20914265 105 + 23771897 UUUAAUCAAAUUCACAUUAACAACAGCAAAGCCUGCAACUGCAACAGC---AA-GACUGCAACACCAACAGCGGCAAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGA ..............................((.((((.((((....((---(.-...)))..........((.((....)).)).....-....)))).))))))..... ( -21.90) >DroSec_CAF1 170411 105 + 1 UUUAAUCAAAUUCACAUUAACAACAGCAAAGCCUGCAACUGCAACAGC---AA-GACUGCAACACCAACUGCGGCAAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGA ..............................((.((((.((((....((---(.-..(((((........)))))....))).((.....-..)))))).))))))..... ( -23.90) >DroSim_CAF1 167378 105 + 1 UUUAAUCAAAUUCACAUUAACAACAGCACAGCCUGCAACUGCAACAGC---GA-GACUGCAACACCAACUGCGGCGAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGA ............................((..(((((.((((....((---(.-(.(((((........))))))...))).((.....-..)))))).)))))...)). ( -24.20) >DroWil_CAF1 215663 88 + 1 UUUAAUCAAAUUCACAUUAACAACAGCA------GCAGCAGCCGCAGC---AA--------AAGCCAACU---GCAACUGCAACAAUUUCCAAUGCAGCUGCAAUAAC-- .........................(((------((.((((..((...---..--------..))...))---))...((((...........)))))))))......-- ( -18.10) >DroYak_CAF1 173183 109 + 1 UUUAAUCAAAUUCACAUUAACAAUAGCAAAGGCUGCAGCCUGAACAACUGCAACACCAACUGCAGCAAAGACAGCAAAGGCAGCAAUUU-UAGCGCAGCUGCAGCAAAGA ...............................(((((((((((.....(((((........)))))......))).....((.((.....-..)))).))))))))..... ( -25.80) >consensus UUUAAUCAAAUUCACAUUAACAACAGCAAAGCCUGCAACUGCAACAGC___AA_GACUGCAACACCAACUGCGGCAAAUGCAGCAAUUU_UAGCGCAGCUGCAGCAAUGA ................................(((((.((((...............................((....)).((........)))))).)))))...... (-10.08 = -11.16 + 1.08)


| Location | 20,914,265 – 20,914,370 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -17.84 |
| Energy contribution | -20.40 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20914265 105 - 23771897 UCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUUGCCGCUGUUGGUGUUGCAGUC-UU---GCUGUUGCAGUUGCAGGCUUUGCUGUUGUUAAUGUGAAUUUGAUUAAA (((...(((((((((((.((-..(..((((((..(((((....))))).)))))).-.)---..)).)))))))))))..((..(..........)..))..)))..... ( -31.40) >DroSec_CAF1 170411 105 - 1 UCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUUGCCGCAGUUGGUGUUGCAGUC-UU---GCUGUUGCAGUUGCAGGCUUUGCUGUUGUUAAUGUGAAUUUGAUUAAA (((...((((((((((((..-.((((((.((....)).))))))...)).((((..-..---.)))).))))))))))..((..(..........)..))..)))..... ( -32.00) >DroSim_CAF1 167378 105 - 1 UCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUCGCCGCAGUUGGUGUUGCAGUC-UC---GCUGUUGCAGUUGCAGGCUGUGCUGUUGUUAAUGUGAAUUUGAUUAAA ((((.((.(((((.((((((-..(((((.((....)).))))))))))).((((((-((---((((...))).)).)))))))))))).))....))))........... ( -33.30) >DroWil_CAF1 215663 88 - 1 --GUUAUUGCAGCUGCAUUGGAAAUUGUUGCAGUUGC---AGUUGGCUU--------UU---GCUGCGGCUGCUGC------UGCUGUUGUUAAUGUGAAUUUGAUUAAA --.....((((((((((...........)))))))))---)((((((..--------..---((.(((((....))------))).)).))))))............... ( -28.40) >DroYak_CAF1 173183 109 - 1 UCUUUGCUGCAGCUGCGCUA-AAAUUGCUGCCUUUGCUGUCUUUGCUGCAGUUGGUGUUGCAGUUGUUCAGGCUGCAGCCUUUGCUAUUGUUAAUGUGAAUUUGAUUAAA .....(((((((((......-.((((((.(((...(((((.......))))).)))...)))))).....))))))))).((..(..........)..)).......... ( -31.90) >consensus UCAUUGCUGCAGCUGCGCUA_AAAUUGCUGCAUUUGCCGCAGUUGGUGUUGCAGUC_UU___GCUGUUGCAGUUGCAGGCUUUGCUGUUGUUAAUGUGAAUUUGAUUAAA ......((((((((((((....((((((.((....)).))))))...)).(((((.......))))).))))))))))................................ (-17.84 = -20.40 + 2.56)


| Location | 20,914,295 – 20,914,385 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -14.32 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20914295 90 + 23771897 GCCUGCAACUGCAACAGCAAGACUGCAACACCAACAGCGGCAAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGAAGGCAAGUCACAACU ((((.((..(((....(((....)))..........(((((...(((.((.....-..)))))))))).))).)).))))........... ( -24.50) >DroSec_CAF1 170441 90 + 1 GCCUGCAACUGCAACAGCAAGACUGCAACACCAACUGCGGCAAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGAAGGCAAGUCACAACU ((((.((..(((..((((..(.(((((........))))))...(((.((.....-..)))))))))..))).)).))))........... ( -26.20) >DroSim_CAF1 167408 90 + 1 GCCUGCAACUGCAACAGCGAGACUGCAACACCAACUGCGGCGAAUGCAGCAAUUU-UAGCGCAGCUGCAGCAAUGAAGGCAAGUCACAACU ((((.((..(((..((((..(.(((((........))))))...(((.((.....-..)))))))))..))).)).))))........... ( -26.20) >DroWil_CAF1 215691 75 + 1 ----GCAGCAGCCGCAGCAA-------AAGCCAACU---GCAACUGCAACAAUUUCCAAUGCAGCUGCAAUAAC--UGGCAACUCGCAACU ----((.(((((.(((((..-------..))....(---((....)))...........))).)))))......--.(....)..)).... ( -19.40) >consensus GCCUGCAACUGCAACAGCAAGACUGCAACACCAACUGCGGCAAAUGCAGCAAUUU_UAGCGCAGCUGCAGCAAUGAAGGCAAGUCACAACU ..(((((.((((....(((...(((((........)))))....))).((........)))))).)))))..................... (-14.32 = -15.70 + 1.38)


| Location | 20,914,295 – 20,914,385 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.75 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20914295 90 - 23771897 AGUUGUGACUUGCCUUCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUUGCCGCUGUUGGUGUUGCAGUCUUGCUGUUGCAGUUGCAGGC .....................(((((((((((.((-..(..((((((..(((((....))))).))))))..)..)).))))))))))).. ( -30.60) >DroSec_CAF1 170441 90 - 1 AGUUGUGACUUGCCUUCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUUGCCGCAGUUGGUGUUGCAGUCUUGCUGUUGCAGUUGCAGGC .....................((((((((((((..-.((((((.((....)).))))))...)).((((.....)))).)))))))))).. ( -31.20) >DroSim_CAF1 167408 90 - 1 AGUUGUGACUUGCCUUCAUUGCUGCAGCUGCGCUA-AAAUUGCUGCAUUCGCCGCAGUUGGUGUUGCAGUCUCGCUGUUGCAGUUGCAGGC ...........((((.((..((((((((.(((...-.....((((((..(((((....))))).))))))..))).)))))))))).)))) ( -33.90) >DroWil_CAF1 215691 75 - 1 AGUUGCGAGUUGCCA--GUUAUUGCAGCUGCAUUGGAAAUUGUUGCAGUUGC---AGUUGGCUU-------UUGCUGCGGCUGCUGC---- ((((((.(((.((((--...((((((((((((...........)))))))))---)))))))..-------..))))))))).....---- ( -31.90) >consensus AGUUGUGACUUGCCUUCAUUGCUGCAGCUGCGCUA_AAAUUGCUGCAUUUGCCGCAGUUGGUGUUGCAGUCUUGCUGUUGCAGUUGCAGGC .....................((((((((((......((((((.((....)).))))))((((.........))))...)))))))))).. (-21.50 = -22.75 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:50 2006