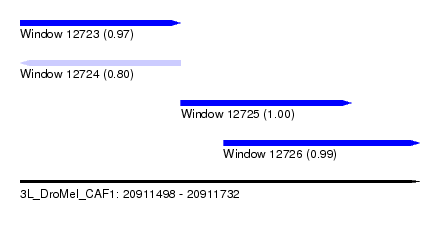
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,911,498 – 20,911,732 |
| Length | 234 |
| Max. P | 0.999783 |

| Location | 20,911,498 – 20,911,592 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20911498 94 + 23771897 CAUUUUGGUGGGAACUAAACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAAGGAAAAACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...(((((((....).......((((.(((((((((((........))))))).(......).)))).))))))))))..(((.......))). ( -24.30) >DroSec_CAF1 168294 92 + 1 CAUUUUGGUGGGAAAUAAACUGGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGA--AACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...((((((((......(((((...((((.((((((((........)))))))))))--).)))))....))))))))..(((.......))). ( -30.70) >DroSim_CAF1 164645 92 + 1 CAUUUUGGUGGGAACUAAACUGGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGA--AACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...((((((((......(((((...((((.((((((((........)))))))))))--).)))))....))))))))..(((.......))). ( -30.70) >DroEre_CAF1 168251 92 + 1 CAUUUUGGUGGGAACUAAACUUGGCUGCCACUCCAAUUGAAACUCGAAUUGGAGCGA--AACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...((((((....)))))).(((((.(((..(((((((........)))))))((((--.....))))))).)))))...(((.......))). ( -28.20) >DroYak_CAF1 170292 92 + 1 CAUUUUGGUGGGAACUAAACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGA--AACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...((((((((......((((.(..((((.((((((((........)))))))))))--).)))))....))))))))..(((.......))). ( -27.80) >consensus CAUUUUGGUGGGAACUAAACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGA__AACAGUUGCGGCCGCCAAAUUGACUUCUCGUGUCG ...((((((((......((((.(......(((((((((........)))))))))......)))))....))))))))..(((.......))). (-24.20 = -24.80 + 0.60)

| Location | 20,911,498 – 20,911,592 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20911498 94 - 23771897 CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUUUUUCCUUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCAAAAUG .(((.......)))..(((((.((..(((((((((((..((.((((((.(.....)))))))))..)))))..))))..))..)).)))))... ( -22.00) >DroSec_CAF1 168294 92 - 1 CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUU--UCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUUUAUUUCCCACCAAAAUG .(((.......)))..(((((.((....(((((((--(((((((((((.(.....))))))))).))))))..))))......)).)))))... ( -25.90) >DroSim_CAF1 164645 92 - 1 CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUU--UCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUUUAGUUCCCACCAAAAUG .(((.......)))..(((((.((..(((((((((--(((((((((((.(.....))))))))).))))))..))))..))..)).)))))... ( -27.40) >DroEre_CAF1 168251 92 - 1 CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUU--UCGCUCCAAUUCGAGUUUCAAUUGGAGUGGCAGCCAAGUUUAGUUCCCACCAAAAUG ......(.(((.(.((((((((.(((((....)).--..(((((((((.(.....))))))))))))).)))))))).).))).)......... ( -28.90) >DroYak_CAF1 170292 92 - 1 CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUU--UCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCAAAAUG .(((.......)))..(((((.((..(((((((((--(((((((((((.(.....))))))))).))))))..))))..))..)).)))))... ( -27.40) >consensus CGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUU__UCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCAAAAUG .(((.......)))..(((((.((..((((((.....(((((((((((.(.....)))))))))).)).....))))..))..)).)))))... (-21.20 = -21.52 + 0.32)


| Location | 20,911,592 – 20,911,692 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 97.20 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20911592 100 + 23771897 ACUGUUGCAUGCCACACCCAGGCAGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGU .((((.((.((((.......)))))))))).....(((.....((((((((((((....(((.(((....))).)))....))))))))))))....))) ( -37.90) >DroSec_CAF1 168386 100 + 1 ACUGUUGCAUGCCACACCCAGGCAGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGU .((((.((.((((.......)))))))))).....(((.....((((((((((((....(((.(((....))).)))....))))))))))))....))) ( -37.90) >DroSim_CAF1 164737 100 + 1 ACUGUUGCAUGCCACACCCAGGCAGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGU .((((.((.((((.......)))))))))).....(((.....((((((((((((....(((.(((....))).)))....))))))))))))....))) ( -37.90) >DroEre_CAF1 168343 99 + 1 ACUGUUGCAUGCCACACCCAGGCUGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCA-UUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCCAUGGU .((((.(((.(((.......)))))))))).....(((.....((((((((((((.(-((((.(((....))).))).)).))))))))))))....))) ( -38.40) >DroYak_CAF1 170384 99 + 1 GCUGUUGCAUGCCACACCCAGGCUGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCA-UUUGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAUUGGU .((((.(((.(((.......)))))))))).....(((.....((((((((((((..-..(((.............)))..))))))))))))....))) ( -38.42) >consensus ACUGUUGCAUGCCACACCCAGGCAGCGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGU .((((.((.((((.......)))))))))).....(((.....(((((((((((((.....)).....((.....)).....)))))))))))....))) (-35.72 = -36.12 + 0.40)


| Location | 20,911,617 – 20,911,732 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.87 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -36.16 |
| Energy contribution | -36.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20911617 115 + 23771897 CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGUAUCGGUAUCUGGUUUAUAUUCUGAACAGGCACAGGCGGAU (((((((((((((.....((((((((((((....(((.(((....))).)))....))))))))))))....))...)))))))).(((((.....)))))........)))... ( -39.10) >DroSec_CAF1 168411 115 + 1 CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGUAUCGGUAUCUGGUUUAUAUGCUGUACAGGCACAGGCGGAU (((.(....)((((((((((((((((((((....(((.(((....))).)))....))))))))))((((..((((....))))))))......))))).)))))....)))... ( -41.30) >DroSim_CAF1 164762 115 + 1 CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGUAUCGGUAUCUGGUUUAUAUGCUGUACAGGCACAGGCGGAU (((.(....)((((((((((((((((((((....(((.(((....))).)))....))))))))))((((..((((....))))))))......))))).)))))....)))... ( -41.30) >DroEre_CAF1 168368 108 + 1 CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCA-UUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCCAUGGUAUCGGUAUCUGGCUUAUAUGCUGCAC------UGGCCAAU .((((((((((((.....((((((((((((.(-((((.(((....))).))).)).))))))))))))....))...))))))))(((......)))))..------........ ( -38.60) >DroYak_CAF1 170409 109 + 1 CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCA-UUUGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAUUGGUAUCGGUAUCUGGUUUAUAUAGUGCAUA-----UGGCGGAU ..(((((((((.(..(..((((((((((((..-..(((.............)))..))))))))))))..)..)...)))))))))..........(((...-----..)))... ( -39.32) >consensus CGCAGAUAUCGCCUGACAGCCGGCUAUCCGCAGUUCGCAAUCCCCAUUCCGGCAUCCGGAUAGCUGGCCAAUGGUAUCGGUAUCUGGUUUAUAUGCUGCACAGGCACAGGCGGAU ..(((((((((((.....(((((((((((((.....)).....((.....)).....)))))))))))....))...)))))))))............................. (-36.16 = -36.16 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:46 2006