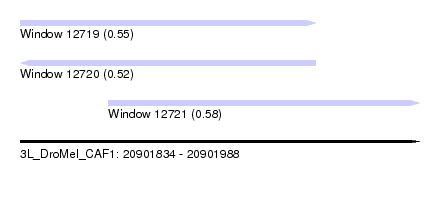
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,901,834 – 20,901,988 |
| Length | 154 |
| Max. P | 0.579798 |

| Location | 20,901,834 – 20,901,948 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -16.37 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20901834 114 + 23771897 ------CUGAUGUACUAAUGGUAAUCAUUAUUUCUGUUUCAUUUAUUUUGGUUUUAGCGUCUAAAGCAGUCUUUUUGCCAUUUACAUUUGCAAAAGACGACCUAUCCCUAAAACUAAAAG ------.((((.(((.....)))))))..................((((((((((((.(..((.....(((((((.((...........)))))))))....))..))))))))))))). ( -20.30) >DroSec_CAF1 158311 114 + 1 ------CUGAUGAACUAAUGGUAAUCAUUAUAUCUGUUUCAUUUAUUUUGGUUUUAGGUUCUAAAGGAGUCUUUUUGGCAUUUACAUUUGCAAAAGACGUCCGAUCCCUAAAACUAAAAG ------..((((((.....((((.......))))...))))))..(((((((((((((.......(((((((((...(((........))).)))))).)))....))))))))))))). ( -29.10) >DroSim_CAF1 154805 114 + 1 ------CUGAUGAACUAAUGGGAAUCAUUAUAUCUGUUUCAUUUAUUUUGGUUUUAGGUUCUAAAAGAGUCUUUUUGGCAUUUACAUUGGCAAAAGACGUCCGAUCCCUAAAACUAAAAG ------..((((...(((((.....)))))))))...........(((((((((((((.((.....))(((((((.(.((.......)).))))))))........))))))))))))). ( -24.80) >DroEre_CAF1 158138 120 + 1 UCAAAUCUGAUUCACUUAUGGUAAUCAUUAUAUCUGAGGUAUUUAUUUUGGUUUUAGCUUCUAAGCCACCCCUUUUACCAUUUACAUGUGCAAAGGACGACCUAUCCCUAAAACUAAAAG ..........((((..((((((....))))))..)))).......((((((((((((((....)))....(((((...(((......))).)))))............))))))))))). ( -18.00) >consensus ______CUGAUGAACUAAUGGUAAUCAUUAUAUCUGUUUCAUUUAUUUUGGUUUUAGCUUCUAAAGCAGUCUUUUUGCCAUUUACAUUUGCAAAAGACGACCGAUCCCUAAAACUAAAAG .......((((...(....)...))))..................(((((((((((((..........(((((((..(((........))))))))))........))))))))))))). (-16.37 = -17.75 + 1.37)

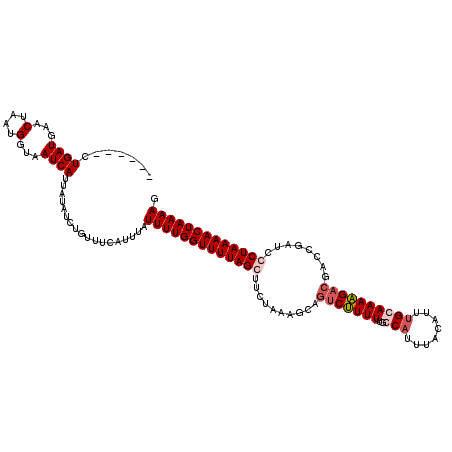
| Location | 20,901,834 – 20,901,948 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -14.90 |
| Energy contribution | -16.77 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20901834 114 - 23771897 CUUUUAGUUUUAGGGAUAGGUCGUCUUUUGCAAAUGUAAAUGGCAAAAAGACUGCUUUAGACGCUAAAACCAAAAUAAAUGAAACAGAAAUAAUGAUUACCAUUAGUACAUCAG------ .((((.(((((((.(..((((.(((((((((...........)).))))))).))))....).))))))).))))..................(((((((.....))).)))).------ ( -21.00) >DroSec_CAF1 158311 114 - 1 CUUUUAGUUUUAGGGAUCGGACGUCUUUUGCAAAUGUAAAUGCCAAAAAGACUCCUUUAGAACCUAAAACCAAAAUAAAUGAAACAGAUAUAAUGAUUACCAUUAGUUCAUCAG------ .((((.((((((((.(..(((.((((((((((........)))..))))))))))..)....)))))))).))))...........((..(((((.....)))))..)).....------ ( -24.30) >DroSim_CAF1 154805 114 - 1 CUUUUAGUUUUAGGGAUCGGACGUCUUUUGCCAAUGUAAAUGCCAAAAAGACUCUUUUAGAACCUAAAACCAAAAUAAAUGAAACAGAUAUAAUGAUUCCCAUUAGUUCAUCAG------ .((((.((((((((.(..(((.(((((((....((....))....))))))))))..)....)))))))).))))...........((..(((((.....)))))..)).....------ ( -20.90) >DroEre_CAF1 158138 120 - 1 CUUUUAGUUUUAGGGAUAGGUCGUCCUUUGCACAUGUAAAUGGUAAAAGGGGUGGCUUAGAAGCUAAAACCAAAAUAAAUACCUCAGAUAUAAUGAUUACCAUAAGUGAAUCAGAUUUGA .((((.(((((((...((((((..(((((...(((....)))....)))))..))))))....))))))).))))........((((((....(((((((.....))).)))).)))))) ( -29.20) >consensus CUUUUAGUUUUAGGGAUAGGACGUCUUUUGCAAAUGUAAAUGCCAAAAAGACUCCUUUAGAACCUAAAACCAAAAUAAAUGAAACAGAUAUAAUGAUUACCAUUAGUUCAUCAG______ .((((.((((((((..(((((((((((((....((....))....))))))).))))))...)))))))).))))...........(((.(((((.....)))))....)))........ (-14.90 = -16.77 + 1.88)


| Location | 20,901,868 – 20,901,988 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -15.07 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20901868 120 + 23771897 AUUUAUUUUGGUUUUAGCGUCUAAAGCAGUCUUUUUGCCAUUUACAUUUGCAAAAGACGACCUAUCCCUAAAACUAAAAGCACUUGAUCUUCCAUCCGCGUUUUUGUUUUCCCACUUCUC .....((((((((((((.(..((.....(((((((.((...........)))))))))....))..)))))))))))))((...((......))...))..................... ( -19.20) >DroSec_CAF1 158345 120 + 1 AUUUAUUUUGGUUUUAGGUUCUAAAGGAGUCUUUUUGGCAUUUACAUUUGCAAAAGACGUCCGAUCCCUAAAACUAAAAGCACUUGAUCUUAUAUCCGCAUUUUUGUUUUCCCACUUCUC .....(((((((((((((.......(((((((((...(((........))).)))))).)))....)))))))))))))......................................... ( -26.20) >DroSim_CAF1 154839 120 + 1 AUUUAUUUUGGUUUUAGGUUCUAAAAGAGUCUUUUUGGCAUUUACAUUGGCAAAAGACGUCCGAUCCCUAAAACUAAAAGCACUUGAUCUUCCAUCCGCAUUUUUGUUUUCCCACUUCUC .....(((((((((((((.((.....))(((((((.(.((.......)).))))))))........)))))))))))))((...((......))...))..................... ( -23.80) >DroEre_CAF1 158178 120 + 1 AUUUAUUUUGGUUUUAGCUUCUAAGCCACCCCUUUUACCAUUUACAUGUGCAAAGGACGACCUAUCCCUAAAACUAAAAGCACUUGAUCUGCCAUCCACAUUUUUGUUCUCCGACUUCUC .....((((((((((((((....)))....(((((...(((......))).)))))............)))))))))))(((.......)))............................ ( -14.50) >consensus AUUUAUUUUGGUUUUAGCUUCUAAAGCAGUCUUUUUGCCAUUUACAUUUGCAAAAGACGACCGAUCCCUAAAACUAAAAGCACUUGAUCUUCCAUCCGCAUUUUUGUUUUCCCACUUCUC .....(((((((((((((..........(((((((..(((........))))))))))........)))))))))))))..................(((....)))............. (-15.07 = -16.07 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:41 2006