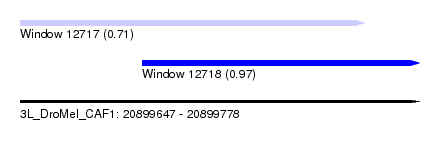
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,899,647 – 20,899,778 |
| Length | 131 |
| Max. P | 0.972589 |

| Location | 20,899,647 – 20,899,760 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -19.38 |
| Energy contribution | -21.66 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20899647 113 + 23771897 AACUUCAAGUAAUUGAAACGAAUGGGUACAUCCUGUAGUCCCCAUAUCUCCAGGUCAAAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACU------AUCCUUGUUCCUACGAGA ...(((((....)))))....(((((.((........)).))))).((((.(((.....((((((((.......))))))))..((((((.....------...)))))))))..)))) ( -32.20) >DroSec_CAF1 156127 113 + 1 AACUUCAAGUAAUUGAAACGAAUGGGUACAUCCUGCAGUCCCCAUAUCUCCAGUUCAAAGCUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACU------AUCCUUGUUCCUACGAGA ...(((((....)))))....(((((.((........)).))))).((((.........((((((((.......))))))))..((((((.....------...)))))).....)))) ( -32.60) >DroSim_CAF1 152623 113 + 1 AACUUCAAGUAAUUGAAACGAAUGGGUACAUCCUGCAGUCCCCAUAUCUCCAGUUCAAAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACU------AUCCUUGUUUCUACGAGA ...(((((....)))))....(((((.((........)).))))).((((.........((((((((.......))))))))..((((((.....------...)))))).....)))) ( -30.20) >DroEre_CAF1 155940 113 + 1 UACUUUAAAUAAUUGAAACGAAUGGGUACAUCCUGCAGUCCCCAUAUCUCCAGGUCAAAGUUGCUGCACUUAUAGCAGUAGCAUGACAAGAUACA------AUCCUUGUUCCUACGAGA .....................(((((.((........)).))))).((((.(((.....((((((((.......))))))))..((((((.....------...)))))))))..)))) ( -28.50) >DroYak_CAF1 157072 113 + 1 UACUUCAAGUAAUUGAAUCGAAUGGGUACAUCCUGCAGUCCCCAUAUCUCCAGAUCGAAGUUGCUGCAAUUAUAGCAGUAACUUGACAAGAUACU------GUCCUUGUUUCUACGAGA ...(((((....)))))....(((((.((........)).))))).((((.(((...((((((((((.......))))))))))((((((.....------...)))))))))..)))) ( -32.40) >DroPer_CAF1 165634 119 + 1 UACUCCAAAAACUGGAAGCGUAUGGGAACAUCUUGCAGACACCAUACCUGAAAGUUGAAUUUGCUGCCCUUGUAGCAAUAGUUGGCCAAGAUGCAUCCAAUAUUUCUGUUUUUAGAUGC ...((.((((((.(((((..(.((((..((((((((((.........)))...((..(..(((((((....)))))))...)..)))))))))..)))))..))))))))))).))... ( -32.00) >consensus AACUUCAAGUAAUUGAAACGAAUGGGUACAUCCUGCAGUCCCCAUAUCUCCAGGUCAAAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACU______AUCCUUGUUCCUACGAGA ...(((((....)))))....(((((.((........)).))))).((((.........((((((((.......))))))))..((((((..............)))))).....)))) (-19.38 = -21.66 + 2.28)

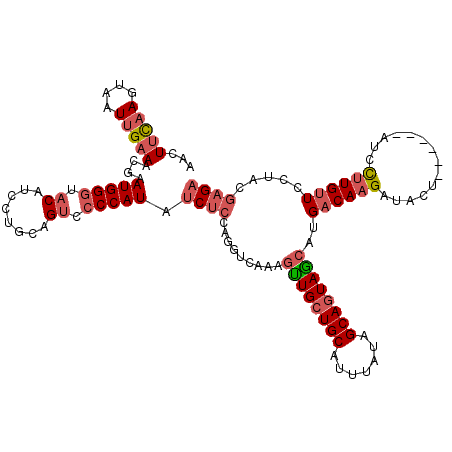

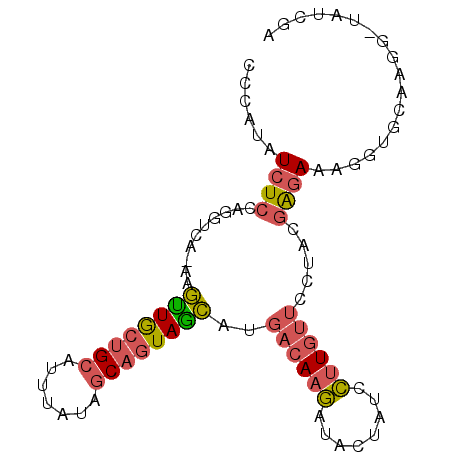
| Location | 20,899,687 – 20,899,778 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20899687 91 + 23771897 CCCAUAUCUCCAGGUCA-AAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACUAUCCUUGUUCCUACGAGAAAUAUGCAAGGUCAUCGA ....(((((....((((-..((((((((.......)))))))).)))).)))))....((((((...............))))))....... ( -23.06) >DroSec_CAF1 156167 90 + 1 CCCAUAUCUCCAGUUCA-AAGCUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACUAUCCUUGUUCCUACGAGAAAGGUGCAAGG-AAUCGA ....(((((...(((..-..((((((((.......))))))))..))).)))))...(((((((.(((.......))).))))))-)..... ( -28.60) >DroSim_CAF1 152663 90 + 1 CCCAUAUCUCCAGUUCA-AAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACUAUCCUUGUUUCUACGAGAAAGGUGCAAGG-AAUCGA ....(((((...(((..-..((((((((.......))))))))..))).)))))...(((((((.(((.......))).))))))-)..... ( -23.80) >DroEre_CAF1 155980 90 + 1 CCCAUAUCUCCAGGUCA-AAGUUGCUGCACUUAUAGCAGUAGCAUGACAAGAUACAAUCCUUGUUCCUACGAGAUAGUUGCAAAG-UUUCGA ....((((((.(((...-..((((((((.......))))))))..((((((........)))))))))..)))))).........-...... ( -24.40) >DroWil_CAF1 204390 91 + 1 ACCAUAUUUGAAAGUUGUAAUCUACCGC-CUUGUAACAAUAGACAGCCACUGUGCAAUCGUUAUAACUAAGGCAAAGGGGUUUCUCUUUUGU .........(((((....(((((...((-((((((((.((.(((((...)))).).)).))))))....))))....)))))...))))).. ( -16.40) >DroYak_CAF1 157112 90 + 1 CCCAUAUCUCCAGAUCG-AAGUUGCUGCAAUUAUAGCAGUAACUUGACAAGAUACUGUCCUUGUUUCUACGAGAUAAGUGCAAAG-UUUCGA ....((((((.(((...-((((((((((.......))))))))))((((((........)))))))))..)))))).........-...... ( -24.80) >consensus CCCAUAUCUCCAGGUCA_AAGUUGCUGCAUUUAUAGCAGUAGCAUGACAAGAUACUAUCCUUGUUCCUACGAGAAAGGUGCAAGG_UAUCGA ......((((..........((((((((.......))))))))..((((((........)))))).....)))).................. (-15.07 = -15.38 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:39 2006