
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
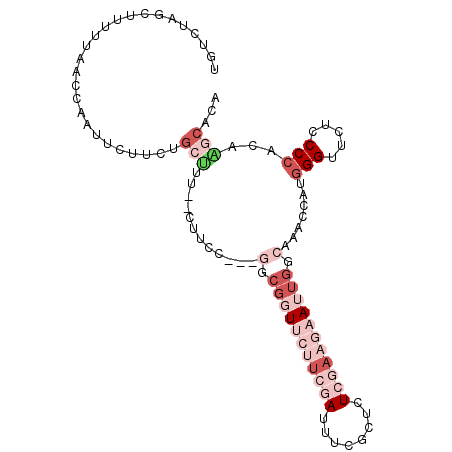
| Location | 20,898,578 – 20,898,680 |
| Length | 102 |
| Max. P | 0.787139 |

| Location | 20,898,578 – 20,898,680 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -7.61 |
| Energy contribution | -11.00 |
| Covariance contribution | 3.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20898578 102 + 23771897 UGUCUAGCUUUUUAUCCAAUUCUUCUGCUUUCGCUUUCGACGGCGUUUCUUCGAUUUCGCUCUCGAAGAAUUGGCAAACAAUGGGUUCUCCCCACAAGCACA ......((((.....((((((((((.((....))....((.((((..((...))...)))).)))))))))))).......((((.....)))).))))... ( -27.60) >DroSec_CAF1 155063 96 + 1 UGUCUAGCUUUUUAGCCAAUUCUUCGGCU------UUCGAAGGCGGUUCUUCGAUUUCGCUCUCGAAGAAUUGGCAAACCACAGGCUCUCCCCACAAGCACA ......((((....((((((((((((((.------.(((((((....)))))))....))...))))))))))))........((......))..))))... ( -31.40) >DroSim_CAF1 151535 96 + 1 UGUCUAGCUUUUUAACCAAUUCUUCUGCU------UUCGAAGGCGGUUCUUCGAUUUCGCUCUCGAAGAAUUGGCAAACCACGGGCUCUCCCCACAAGCACA ......((((.....((((((((((.((.------.(((((((....)))))))....))....))))))))))........(((.....)))..))))... ( -28.40) >DroEre_CAF1 154865 99 + 1 UAUCUAGCUUUUUCACAAAUUCUUCUGCCUUAACUUCC---GGCGGUCGUUCCAUUUGGCCCUCAACAAAUUGGCAAACAAUGGGUUCUCCCCAUAGGCACA ......((((...............((((.........---((.(((((.......))))))).........))))....(((((.....)))))))))... ( -20.47) >DroYak_CAF1 156000 99 + 1 UUUCUACCUUUUUUACCAAUUCGUCUGCUUUGACUUCC---UGCGGUCGUUCUAUUUCACUCUCAAAAAAUUGGCAAACCAUGGGUUCUCCCCAUAUACACA ...............((((((((.((((..........---.)))).))...................))))))......(((((.....)))))....... ( -14.20) >DroPer_CAF1 164484 93 + 1 UCUCCGGC------UCCGACUCGACUACUUCCUCGUCU---UCGGGUUCCGCGAUUUCCUCUUCGAUGAAAAGGCAAACUGUGGGAUUUCCCCAGCGACAGA ......((------(((((..(((........)))...---))))((((((((.((((((.(((...))).))).))).))))))))......)))...... ( -21.90) >consensus UGUCUAGCUUUUUAACCAAUUCUUCUGCUUU__CUUCC___GGCGGUUCUUCGAUUUCGCUCUCGAAGAAUUGGCAAACCAUGGGUUCUCCCCACAAGCACA ..........................(((............(.(((((((((((........))))))))))).).......(((.....)))...)))... ( -7.61 = -11.00 + 3.39)

| Location | 20,898,578 – 20,898,680 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20898578 102 - 23771897 UGUGCUUGUGGGGAGAACCCAUUGUUUGCCAAUUCUUCGAGAGCGAAAUCGAAGAAACGCCGUCGAAAGCGAAAGCAGAAGAAUUGGAUAAAAAGCUAGACA ...(((((((((.....)))))..((((((((((((((((..(((............)))..))....((....)).)))))))))).))))))))...... ( -33.90) >DroSec_CAF1 155063 96 - 1 UGUGCUUGUGGGGAGAGCCUGUGGUUUGCCAAUUCUUCGAGAGCGAAAUCGAAGAACCGCCUUCGAA------AGCCGAAGAAUUGGCUAAAAAGCUAGACA ...(((((..((.....))..).....((((((((((((...((....((((((......)))))).------.))))))))))))))....))))...... ( -33.10) >DroSim_CAF1 151535 96 - 1 UGUGCUUGUGGGGAGAGCCCGUGGUUUGCCAAUUCUUCGAGAGCGAAAUCGAAGAACCGCCUUCGAA------AGCAGAAGAAUUGGUUAAAAAGCUAGACA ...(((..((((.....))))..)...(((((((((((....((....((((((......)))))).------.)).)))))))))))......))...... ( -31.20) >DroEre_CAF1 154865 99 - 1 UGUGCCUAUGGGGAGAACCCAUUGUUUGCCAAUUUGUUGAGGGCCAAAUGGAACGACCGCC---GGAAGUUAAGGCAGAAGAAUUUGUGAAAAAGCUAGAUA ...((..(((((.....)))))..((((((..........(..((....))..)......(---....)....))))))...............))...... ( -19.70) >DroYak_CAF1 156000 99 - 1 UGUGUAUAUGGGGAGAACCCAUGGUUUGCCAAUUUUUUGAGAGUGAAAUAGAACGACCGCA---GGAAGUCAAAGCAGACGAAUUGGUAAAAAAGGUAGAAA ......((((((.....)))))).(((((((((((((((...((........))(((....---....)))....)))).)))))))))))........... ( -23.00) >DroPer_CAF1 164484 93 - 1 UCUGUCGCUGGGGAAAUCCCACAGUUUGCCUUUUCAUCGAAGAGGAAAUCGCGGAACCCGA---AGACGAGGAAGUAGUCGAGUCGGA------GCCGGAGA ......(((((((....))).))))...(((((((...)))))))...((.(((...((((---.(((.........)))...)))).------.))))).. ( -29.70) >consensus UGUGCUUGUGGGGAGAACCCAUGGUUUGCCAAUUCUUCGAGAGCGAAAUCGAAGAACCGCC___GAAAG__AAAGCAGAAGAAUUGGUUAAAAAGCUAGACA ..((((..((((.....))))...........((((((((........)))))))).................))))......(((((......)))))... (-10.70 = -11.23 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:37 2006