
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,892,491 – 20,892,691 |
| Length | 200 |
| Max. P | 0.996546 |

| Location | 20,892,491 – 20,892,611 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -28.76 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20892491 120 - 23771897 AAAUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCAGAGUUCGCAUAAAUCAUUUCUGAUCAAAGCCUUCGGACUUCUGGAAACAUGUGGUUUUCCCGAU ...(((((((......)))..((..((((((((((.((((...(((((.....)))).)...))))..((((....)))).......))))))))))..))....))))........... ( -27.80) >DroSec_CAF1 149135 120 - 1 AAGUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCAGAGUUCGCAUAAAUCAUUUCUGAUCAAAGCCUUCGGACUUCUGGAAACAUGUGGUUUUCCUGCU .....((((.......(((((((..((((((((((.((((...(((((.....)))).)...))))..((((....)))).......))))))))))..))....)).)))...)))).. ( -30.50) >DroSim_CAF1 145605 120 - 1 AAGUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCAGAGUUCGCAUAAAUCAUUUCUGAUCAACGCCUUCGGACUUCUGGAAACAUGUGGUUUUCCUGCU .....((((.......(((((((..((((((((((..((((......(((((.......)))))....((((....)))).))))..))))))))))..))....)).)))...)))).. ( -31.20) >DroEre_CAF1 149010 120 - 1 AAAUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCACAGUUCGCAUAAGUCAUUUUUGAUUAGCUCCUUCGGACUUCUGGAAAUAUGCGGUUUUCCUGAU .....((((.......(((((((..((((((((((...((((.....(((((.......)))))....((((....))))))))...))))))))))..))....)).)))...)))).. ( -33.10) >DroYak_CAF1 150004 120 - 1 AAGUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCACAGUUCGCAUAAAUCAUUUCUGAUCAAGGACUUCGGACUUCUGGAAAUAUGUGGUUUUCCUGAU .....((((.(((((((....((..((((((((((.......((((.(((((.......)))))....((((....))))..)))).))))))))))..))..)))))))....)))).. ( -31.50) >consensus AAGUACAGGUUUAUAUACCCAUCUGGAAGUUUGAAAAUGUUAUCCUGGCGAAACCAGAGUUCGCAUAAAUCAUUUCUGAUCAAAGCCUUCGGACUUCUGGAAACAUGUGGUUUUCCUGAU .....((((.......(((((((..((((((((((............(((((.......)))))....((((....)))).......))))))))))..))....)).)))...)))).. (-28.76 = -28.80 + 0.04)


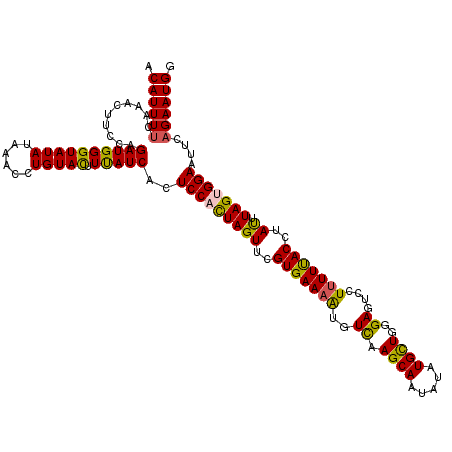
| Location | 20,892,571 – 20,892,691 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20892571 120 + 23771897 ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUAUUUUAUCACUCCACUAGUUCGUGAAAGUGUCAAGCAAUAUAUGCUGGGAGUCCUAUUUACCUAUUUAGUGGAAUUCAGAAUGG .((((((.((..(((((((((((((.(((..............((((.(((......)))..))))((.((((.....))))..))....))).))))))))...))))))).)))))). ( -25.00) >DroSec_CAF1 149215 120 + 1 ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUACUUUAUCACUCCACUAGUUCGUGAAAAUGUCAAGCAAUAUAUGCUGGGAGUCCUUUUUACCUACUUAGUGGAAUUCAGAAUGG .((((((..........((((((((((......))))).)))))..(((((((((..(((((((.(((.((((.....))))..))..).)))))))..)).)))))))....)))))). ( -28.30) >DroSim_CAF1 145685 120 + 1 ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUACUUUAUCACUCCACUAGUUCGUGAAAAUGUCAAGCAAUAUAUGCUGGGAGUCCUUUUUACCUACUUAGUGGAAUUCAGAAUGG .((((((..........((((((((((......))))).)))))..(((((((((..(((((((.(((.((((.....))))..))..).)))))))..)).)))))))....)))))). ( -28.30) >DroEre_CAF1 149090 120 + 1 ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUAUUUCAUCACUCCUCUAGUUCGUGAAAACGUCAAGCAAUAUAUGCUGGGAGUACUUUUCACCUAUUUAGUGGAAUUCAGAAUGG .((((((.((..(((((((((((.(((((....)))))))))))((((((......(((....)))...((((.....)))))))))).................))))))).)))))). ( -27.70) >DroYak_CAF1 150084 120 + 1 ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUACUUUAUCACUCCCUUAGUUCGUGAAAAUGUUAAGCAAUAUAUGUUGGGAGUACUUUUUACCUAUUUAGUGGAGUUCCGAAUGG .(((((...((((((((((((((((((((....)))).......((((((......(((....)))...((((.....))))))))))......))))))))).).))))))..))))). ( -28.40) >consensus ACAUUUUCAAACUUCCAGAUGGGUAUAUAAACCUGUACUUUAUCACUCCACUAGUUCGUGAAAAUGUCAAGCAAUAUAUGCUGGGAGUCCUUUUUACCUAUUUAGUGGAAUUCAGAAUGG .((((((..........((((((((((......))))).)))))..(((((((((..(((((((..((.((((.....))))..))....)))))))..)).)))))))....)))))). (-24.04 = -23.40 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:34 2006