
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,892,291 – 20,892,451 |
| Length | 160 |
| Max. P | 0.961699 |

| Location | 20,892,291 – 20,892,411 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20892291 120 + 23771897 AAUAUAAUGAUCUCAUCCUAUAUACCAUAUGAGAAAAAUGAAUACGAUCAGAGAGAACCUUGGUUCGGAACUCAGUUCAAAACCAUCUCUAUGCCCCCAAAAUGGCAAACUAUGUGGGAA ...............(((((((((...........................(((((....(((((..((((...))))..)))))))))).((((........))))...))))))))). ( -27.00) >DroSec_CAF1 148935 120 + 1 AAUAUAAUGAUCUCAUCCUAUAUACCAUAUGAGAAAAAUGAGUACGACCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCCAUGCCCUCAAAAUGGCAAACCAUGUGGGAA .((((((((....)))..))))).(((((((.(((...(((((.(((((((........)))).)))..))))).))).............((((........))))...)))))))... ( -25.50) >DroSim_CAF1 145405 120 + 1 AAUAUAAUGAUCUCAUCCUAUAUACCAUAUGAGAAAAAUGAGUACGAUCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCCAUGCCCUCAAAAUGGCAAACAAUGUGGGAA ...............((((((((.......((((....(((......)))..........((((((.(((.....))).))))))))))..((((........))))....)))))))). ( -24.30) >DroEre_CAF1 148810 120 + 1 AAUAUAAUGAUCUCAUCUUAUAUGCCAUAUGAAGAGAAUGAAUACGACCAGAAGGAGCCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUUAAAGUGGCAAACUAUGUGGGAA .((((((.(((...))))))))).(((((((.(((((..........((....)).....((((((.(((.....))).))))))))))).(((((.....).))))...)))))))... ( -26.70) >DroYak_CAF1 149804 120 + 1 AAUAUAAUGAUCUCAUCCUAUAUACCAUAUGAAGAAAAUGAUUACGACCAGAAAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUCAAAAUGGCAAACUAUGUGGGAA ...............(((((((((.............................(((....((((((.(((.....))).))))))..))).((((........))))...))))))))). ( -21.70) >consensus AAUAUAAUGAUCUCAUCCUAUAUACCAUAUGAGAAAAAUGAAUACGACCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUCAAAAUGGCAAACUAUGUGGGAA .((((((((....)))..))))).(((((((.....................((((....((((((.(((.....))).))))))))))..((((........))))...)))))))... (-20.78 = -21.30 + 0.52)

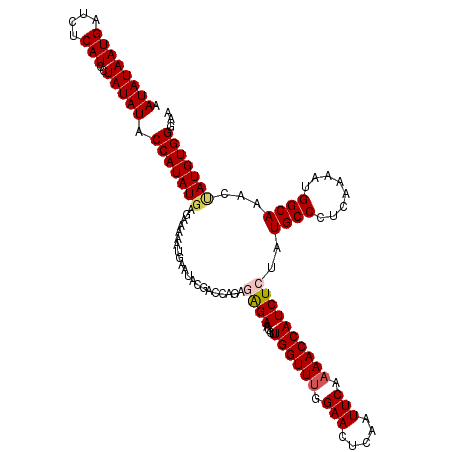
| Location | 20,892,331 – 20,892,451 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20892331 120 + 23771897 AAUACGAUCAGAGAGAACCUUGGUUCGGAACUCAGUUCAAAACCAUCUCUAUGCCCCCAAAAUGGCAAACUAUGUGGGAACAGUCAGCGACUAUAAAAGAGUUGCACUAUCCGCAACCAA ...........(((((....(((((..((((...))))..)))))))))).((((........)))).....((((((...(((..((((((.......))))))))).))))))..... ( -30.70) >DroSec_CAF1 148975 120 + 1 AGUACGACCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCCAUGCCCUCAAAAUGGCAAACCAUGUGGGAACAGUCAGCGACAUUAAAGGAGUUGCACUAUCCGCAGCCAA .....((.((..((((....((((((.(((.....))).))))))))))..))...))....((((......((((((...(((((((..(......)..)))).))).)))))))))). ( -27.90) >DroSim_CAF1 145445 120 + 1 AGUACGAUCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCCAUGCCCUCAAAAUGGCAAACAAUGUGGGAAAAGUCAGCGACUUUAAAGGAGUUGCACUAUCCGCAGCCAA .....((.((..((((....((((((.(((.....))).))))))))))..))...))....((((......((((((...(((..(((((((.....)))))))))).)))))))))). ( -29.70) >DroEre_CAF1 148850 120 + 1 AAUACGACCAGAAGGAGCCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUUAAAGUGGCAAACUAUGUGGGAACAGCCAGCGACCUUAAAAGAGUUGCAUUAUCCGCAACCCA .............((((...((((((.(((.....))).)))))).)))).(((((.....).)))).....((((((......).(((((((....)).))))).....)))))..... ( -26.50) >DroYak_CAF1 149844 120 + 1 AUUACGACCAGAAAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUCAAAAUGGCAAACUAUGUGGGAACAGCCAGCGACUUUAAACGAGUUGCAAUAUCCGCAACCGA ...................((((((((((.............((((.....((((........))))......)))).........(((((((.....)))))))....)))).)))))) ( -25.50) >consensus AAUACGACCAGAGAGAACCUUGGUUUGGAACUCAAUUCAAAACCAUCUCUAUGCCCUCAAAAUGGCAAACUAUGUGGGAACAGUCAGCGACUUUAAAAGAGUUGCACUAUCCGCAACCAA ...................((((((((((.............((((.....((((........))))......)))).........(((((((.....)))))))....)))).)))))) (-23.94 = -24.22 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:32 2006