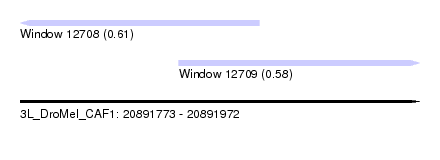
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,891,773 – 20,891,972 |
| Length | 199 |
| Max. P | 0.612201 |

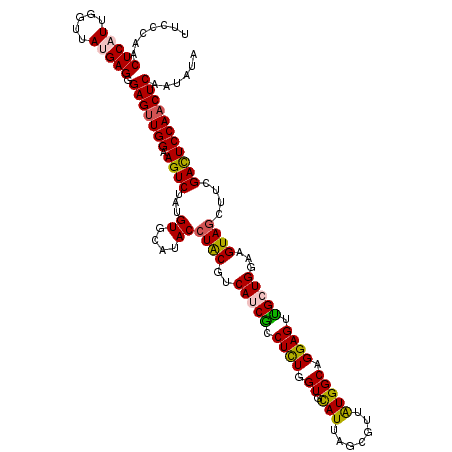
| Location | 20,891,773 – 20,891,892 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.45 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20891773 119 - 23771897 ACAUAGACUUCCAACUCCCUCAUAACCAAUGAGUUGGGAAAUAAACAUAUC-CGGCUUGCUGCGGUAAAAGUUCUUCAUUGGGGGUAGCACCCAAGUAAGCUCCAGCUUGCACACAUCUU .....((((((((((((.............)))))))))........((((-(((....))).))))...))).....(((((.......)))))((((((....))))))......... ( -29.62) >DroSec_CAF1 148417 119 - 1 ACAUAGACUGCCAUCUCCCUCAUAACCAAUGAGUUGGGAAAUAAACAUAUC-CGGCUUGCUGCGGUAAAAGUCCUUCAUUGGGGGUAACACCCAAGUCAUCUCCAGCUUGCACACAUCUU .....((((................(((((((...((((........((((-(((....))).))))....)))))))))))(((.....))).))))...................... ( -25.40) >DroEre_CAF1 148292 119 - 1 ACAUAGACUUCCAACUCCCUCGUAACCAAUGAGUUGGGAAAUAAAAAUAUC-CGGCUUGCUGCGAUAGAAGUUCUUCAUUGGGGGUAAUACCCAAGUUAGCUCCAGCUUGCACACGUCUU ....(((((((((((((.............)))))))))............-..((..((((.((..(((....))).(((((.......)))))......))))))..))....)))). ( -31.72) >DroYak_CAF1 149294 111 - 1 ACAUAGACUUCCAACUCCCUCAUAACCAAGGAGUUGGGAAAUAAAAAUAUCCCGGCUUGCUGCGAUAAAAGUUCUUCAUUGGGGGUAACACCCAAGUCAGCUCCANNNNNN--------- .....(((((((((((((...........))))))))))........((((.(((....))).))))...........(((((.......)))))))).............--------- ( -28.70) >consensus ACAUAGACUUCCAACUCCCUCAUAACCAAUGAGUUGGGAAAUAAAAAUAUC_CGGCUUGCUGCGAUAAAAGUUCUUCAUUGGGGGUAACACCCAAGUCAGCUCCAGCUUGCACACAUCUU .....((((((((((((.............)))))))..........((((.(((....))).)))).))))).....(((((.......)))))......................... (-21.23 = -21.30 + 0.06)

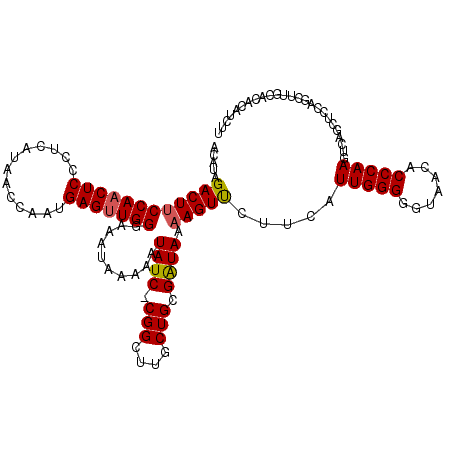
| Location | 20,891,852 – 20,891,972 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -28.55 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20891852 120 + 23771897 UUCCCAACUCAUUGGUUAUGAGGGAGUUGGAAGUCUAUGUGCAUACCUACGUCAUCACCUUUGGUGCAUUAGCGUUGUGGCAGGAGUUGCUGAAAGUAGUUUCGAUUCCAACUCAAUUUA .......(((((.....))))).(((((((((.((....(((.(((..((((...((((...)))).....)))).))))))(((..(((.....)))..))))))))))))))...... ( -34.10) >DroSec_CAF1 148496 120 + 1 UUCCCAACUCAUUGGUUAUGAGGGAGAUGGCAGUCUAUGUGCAUACCUACGUCAUCGCCUCUGGUGUAUUAGCGUUGUGGCAGGAGUUGCUGGAAGUAGUUUCGACUCCAACUCGAUUUA .......(((((.....))))).(((.(((.((((....(((.(((..((((...((((...)))).....)))).))))))(((..(((.....)))..)))))))))).)))...... ( -32.50) >DroEre_CAF1 148371 120 + 1 UUCCCAACUCAUUGGUUACGAGGGAGUUGGAAGUCUAUGUUCCUACCUACGCCAUCGUCUCUGGUGCAUUAGCGUUAUGGCCGGAGUCGGUGGAAGUAGCUUCGACUCCAACUCCAUAUA ...((((....)))).......(((((((((.(((...((....))((((.((((((.(((((((.(((.......)))))))))).))))))..))))....))))))))))))..... ( -45.70) >DroYak_CAF1 149365 120 + 1 UUCCCAACUCCUUGGUUAUGAGGGAGUUGGAAGUCUAUGUUCUUACGUGCGACAUCGUCUCUGGUGCAUUAGCGUUAUGGCAGGAGUUGGUGGAAGUAGCUUCGACUCCAACUCAAUAUA ...((((((((((.......))))))))))..((((((((....))))).)))...(((..(..(((....)))..).)))..((((((((.((((...)))).)..)))))))...... ( -38.70) >consensus UUCCCAACUCAUUGGUUAUGAGGGAGUUGGAAGUCUAUGUGCAUACCUACGUCAUCGCCUCUGGUGCAUUAGCGUUAUGGCAGGAGUUGCUGGAAGUAGCUUCGACUCCAACUCAAUAUA .......(((((.....))))).(((((((.((((...((....))((((..(((((.((((.((.(((.......))))).)))).)))))...))))....)))))))))))...... (-28.55 = -28.68 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:30 2006