
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,887,479 – 20,887,580 |
| Length | 101 |
| Max. P | 0.976070 |

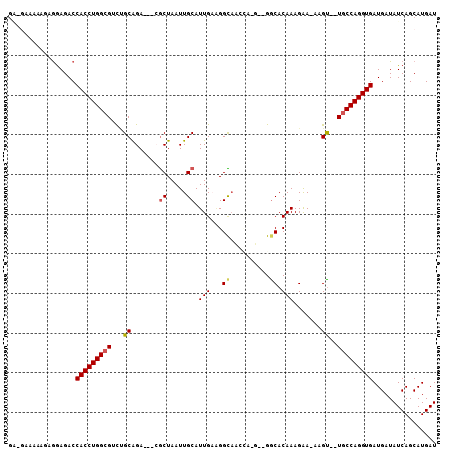
| Location | 20,887,479 – 20,887,580 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
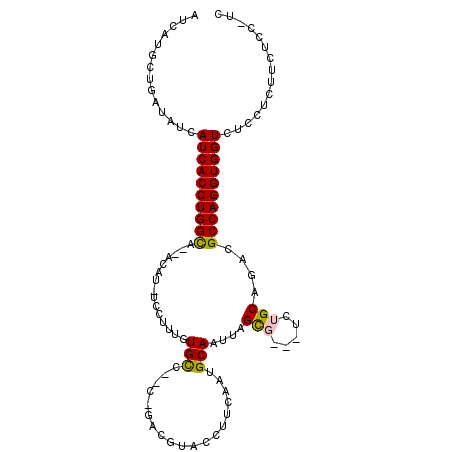
>3L_DroMel_CAF1 20887479 101 + 23771897 GC-GAGAAACUGGAGACCACCUGGCGUCUGCAGACCCCGCUAAUUGCAUUGUGGGUACGCA-G--GGCACAAAGGA-AAGU--UGCCAGGUGAUGAUAUCAGCAUGAU ((-..((....(....)((((((((((((((.(..(((((..........)))))..))))-.--)))((......-..))--.))))))))......)).))..... ( -31.70) >DroVir_CAF1 178783 91 + 1 --------AGAGGAGACCACCUGGCGUCAGCGGA---GGCUAAUUGCAUUGAAGGCAACGC-U--GGCACAAAGAA-AUGU--UGCCAGGUGAUGAUAUCAGCAUGAU --------.(((....)(((((((((((((((..---.(((............)))..)))-)--)))(((.....-.)))--.))))))))......))........ ( -32.40) >DroGri_CAF1 163740 102 + 1 GAUGGAGAAGAGGAGACCACCUGGCGUCUGCAGA---CGCUAAUUGCAUUGAAGGCAACA--GCAGACACAAAGAA-AUGUUUUGCCAGGUGAUGAUAUCAGCAUGAU ((((.......(....)((((((((((((((...---.((.....))......(....).--))))))(((.....-.)))...))))))))....))))........ ( -33.60) >DroWil_CAF1 196278 101 + 1 GA-GAGUAA--AGAGACCACCUGGCGUUUGCAGAGAACACUAAUUGCAUUGAUGGUCCGUGUG--GGCACAAAGGAAAAGU--UACCAGGUGAUGAUAUCAGCAUGAU ..-......--......(((((((....((((.((....))...)))).((((..(((...((--....))..)))...))--)))))))))................ ( -21.50) >DroMoj_CAF1 173482 101 + 1 GACGUAGCAGAGGAGACCACCUGGCGUUCGCGGA---GGCUAAUUGCAUUGAAGGCAACUC-UCAGGCACAAAGAA-AUGC--UGCCAGGUGAUGAUAUCAGCAUGAU ..(((.((.(((....)(((((((((...(((..---..((...(((.((((((....)).-)))))))...))..-.)))--)))))))))......)).))))).. ( -31.50) >DroAna_CAF1 143443 101 + 1 GG-CGAAGCCCGAAGACCACCUGGCGGCCGCAGAGCCCGCUAAUUGCAUUGUGGGUACGCA-G--GGCACAAAGGA-AAGU--UGCCAGGUGAUGAUAUCAGCAUGAU ((-(...))).......(((((((((((..(.(.((((((.....))..((((....))))-)--))).)....).-..))--)))))))))................ ( -34.70) >consensus GA_GAAAAAGAGGAGACCACCUGGCGUCUGCAGA___CGCUAAUUGCAUUGAAGGCAACCA_G__GGCACAAAGAA_AAGU__UGCCAGGUGAUGAUAUCAGCAUGAU .................(((((((((...((.......((.....)).(((...((..........)).))).......))..)))))))))................ (-17.41 = -17.60 + 0.19)


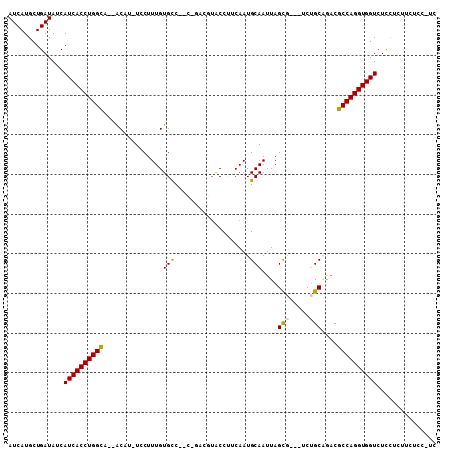
| Location | 20,887,479 – 20,887,580 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20887479 101 - 23771897 AUCAUGCUGAUAUCAUCACCUGGCA--ACUU-UCCUUUGUGCC--C-UGCGUACCCACAAUGCAAUUAGCGGGGUCUGCAGACGCCAGGUGGUCUCCAGUUUCUC-GC .....((((.....((((((((((.--((..-......))...--(-((((.((((....(((.....))))))).)))))..))))))))))...)))).....-.. ( -34.50) >DroVir_CAF1 178783 91 - 1 AUCAUGCUGAUAUCAUCACCUGGCA--ACAU-UUCUUUGUGCC--A-GCGUUGCCUUCAAUGCAAUUAGCC---UCCGCUGACGCCAGGUGGUCUCCUCU-------- ..............((((((((((.--(((.-.....)))...--.-((((((....))))))..(((((.---...))))).)))))))))).......-------- ( -28.20) >DroGri_CAF1 163740 102 - 1 AUCAUGCUGAUAUCAUCACCUGGCAAAACAU-UUCUUUGUGUCUGC--UGUUGCCUUCAAUGCAAUUAGCG---UCUGCAGACGCCAGGUGGUCUCCUCUUCUCCAUC ...(((..((....((((((((((...(((.-.....)))((((((--.(((((.......))))).....---...)))))))))))))))).......))..))). ( -31.60) >DroWil_CAF1 196278 101 - 1 AUCAUGCUGAUAUCAUCACCUGGUA--ACUUUUCCUUUGUGCC--CACACGGACCAUCAAUGCAAUUAGUGUUCUCUGCAAACGCCAGGUGGUCUCU--UUACUC-UC ..............((((((((((.--........((((((..--..)))))).......((((...((....)).))))...))))))))))....--......-.. ( -21.50) >DroMoj_CAF1 173482 101 - 1 AUCAUGCUGAUAUCAUCACCUGGCA--GCAU-UUCUUUGUGCCUGA-GAGUUGCCUUCAAUGCAAUUAGCC---UCCGCGAACGCCAGGUGGUCUCCUCUGCUACGUC .....((.((....((((((((((.--((((-......))))..((-(((((((.......))))))...)---)).......))))))))))....)).))...... ( -29.90) >DroAna_CAF1 143443 101 - 1 AUCAUGCUGAUAUCAUCACCUGGCA--ACUU-UCCUUUGUGCC--C-UGCGUACCCACAAUGCAAUUAGCGGGCUCUGCGGCCGCCAGGUGGUCUUCGGGCUUCG-CC .....((.........((((((((.--....-.....(((...--.-(((((.......)))))....)))((((....))))))))))))(((....)))...)-). ( -32.50) >consensus AUCAUGCUGAUAUCAUCACCUGGCA__ACAU_UCCUUUGUGCC__C_GACGUACCUUCAAUGCAAUUAGCG___UCUGCAGACGCCAGGUGGUCUCCUCUUCUCC_UC ..............((((((((((...............(((...................)))....(((.....)))....))))))))))............... (-18.24 = -18.16 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:26 2006