| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,861,364 – 20,861,461 |
| Length | 97 |
| Max. P | 0.938986 |

| Location | 20,861,364 – 20,861,461 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.79 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
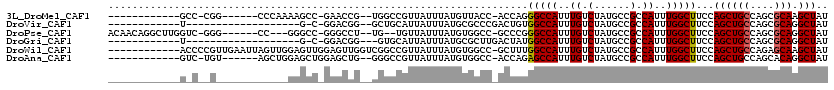
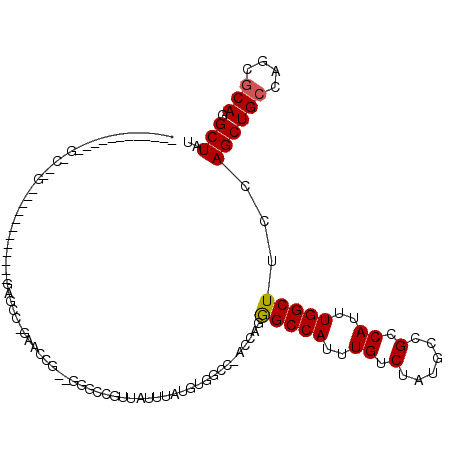
>3L_DroMel_CAF1 20861364 97 + 23771897 AUAGCUUGCGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCCUGGU-GGUAACAUAAAUAACGGCCA--CGGUUC-GGCUUUUGGG------CCG-GGC------------ .......((.(((((..(..((((((.(((.(.(((........(((..((....-))...)))........))).--).))).-))))))..))------)))-)))------------ ( -33.19) >DroVir_CAF1 145480 85 + 1 AUAGCCUGCGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCACAGUCGGGCGCAUAAAUAAUGCAGC--CCGUCC-G-C-------------------A------------ ...(((......))).((.(((.(((....)))(((.((......))))).....(((((((((.....)))).))--))))))-)-)-------------------.------------ ( -32.00) >DroPse_CAF1 131034 104 + 1 AUAGCCUGCGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCCGGGC-GGCCACAUAAAUAACA--CA--AGGCCC-GGCCC---GG------CCC-GACCAAGCCUGUUGU (((((..((((((.((..(((...)))..)).)))).........(((.((((((-((((............--..--.)))).-.))))---))------.))-).....))..))))) ( -38.06) >DroGri_CAF1 132207 84 + 1 AUAGCCUGCGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCAUAGUCAAGCGCAUAAAUAAUGCAC---CCGUCC-G-C-------------------A------------ .........((.(((.(((.((.(((....)))(((.((......)))))....)).)))((((.....))))..---..))).-)-)-------------------.------------ ( -23.00) >DroWil_CAF1 166501 107 + 1 AUAGCUUGCUCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCAAAGC-GGCCACAUAAAUAACGGCCGACCAACUCCAACUCCAACUAAUUCAACGGGGU------------ ..((((.((....))))))(((.(((....)))(((.........(((((.....-)))))...........))).......))).(((((............)))))------------ ( -30.45) >DroAna_CAF1 118771 98 + 1 AUAGCCUGUGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCUCUGGU-GGCCACAUAAAUAACGGCCC--CAGCUCCAGCUCCAGCU------ACA-GAC------------ .....(((((((((.((((((((((((..((.(......).))..)))))((((.-((((...........)))))--))).)))))))))))).------)))-)..------------ ( -45.70) >consensus AUAGCCUGCGCUGGCAGCUGGAAGCCAAAUGGCGGCAUAGACAAAUGGCCAUAGU_GGCCACAUAAAUAACGGCCA__CAGCCC_GGCUC____________C__G_C____________ .((((.(((....)))))))(..(((....)))..).........(((((......)))))........................................................... (-14.82 = -15.10 + 0.28)

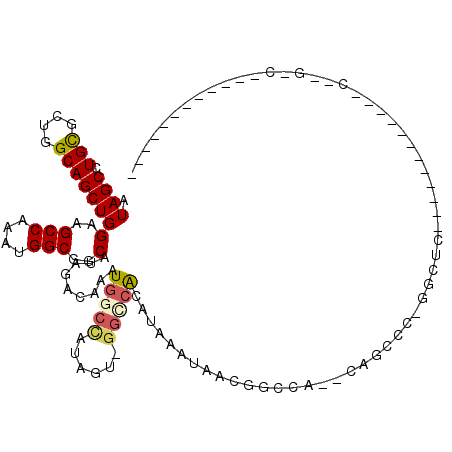
| Location | 20,861,364 – 20,861,461 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.79 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20861364 97 - 23771897 ------------GCC-CGG------CCCAAAAGCC-GAACCG--UGGCCGUUAUUUAUGUUACC-ACCAGGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCGCAAGCUAU ------------...-.((------(....(((((-(((..(--((((....((..(((...((-....))..)))..))....)))))..))))))))......)))(((....))).. ( -30.50) >DroVir_CAF1 145480 85 - 1 ------------U-------------------G-C-GGACGG--GCUGCAUUAUUUAUGCGCCCGACUGUGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCGCAGGCUAU ------------.-------------------(-(-((.(((--((.((((.....))))))))).))))(((.((......)))))(((..(((((........)))))....)))... ( -31.90) >DroPse_CAF1 131034 104 - 1 ACAACAGGCUUGGUC-GGG------CC---GGGCC-GGGCCU--UG--UGUUAUUUAUGUGGCC-GCCCGGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCGCAGGCUAU ......(((.(((.(-((.------((---((((.-.((((.--((--((.....)))).))))-)))))).))....).))).)))(((..(((((........)))))....)))... ( -42.80) >DroGri_CAF1 132207 84 - 1 ------------U-------------------G-C-GGACGG---GUGCAUUAUUUAUGCGCUUGACUAUGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCGCAGGCUAU ------------.-------------------(-(-((((((---((((((.....))))))))).....(((((..((.(......).))..))))))))..((((....))))))... ( -28.50) >DroWil_CAF1 166501 107 - 1 ------------ACCCCGUUGAAUUAGUUGGAGUUGGAGUUGGUCGGCCGUUAUUUAUGUGGCC-GCUUUGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGAGCAAGCUAU ------------.....(((....((((((((((..((..(((.((((...((..((.((((((-.....)))))).))..)).)))))))))..)).))))))))....)))....... ( -36.20) >DroAna_CAF1 118771 98 - 1 ------------GUC-UGU------AGCUGGAGCUGGAGCUG--GGGCCGUUAUUUAUGUGGCC-ACCAGAGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCACAGGCUAU ------------(((-(((------.((((((((((((.(((--((((((.........)))))-.))))(((((..((.(......).))..)))))))))))).)))))))))))... ( -52.40) >consensus ____________G_C__G____________GAGCC_GAACCG__GGGCCGUUAUUUAUGUGGCC_ACCAGGGCCAUUUGUCUAUGCCGCCAUUUGGCUUCCAGCUGCCAGCGCAGGCUAU ......................................................................(((((..((.(......).))..)))))...((((((....))).))).. (-12.27 = -12.30 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:17 2006