
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,860,672 – 20,860,775 |
| Length | 103 |
| Max. P | 0.851720 |

| Location | 20,860,672 – 20,860,775 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
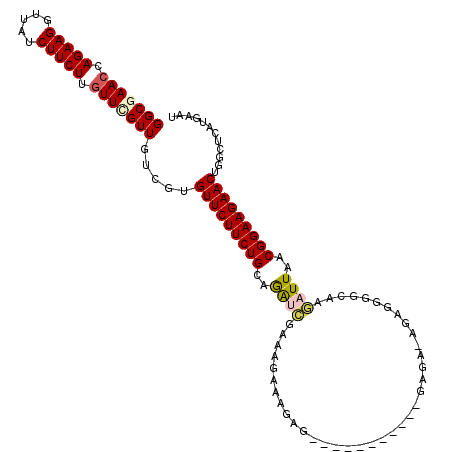
>3L_DroMel_CAF1 20860672 103 + 23771897 GGCGAACCAGAAGGUUAUCUUCUUGUUUGUUGUCGUGUUCUUCUGCAGGUCGAAAGAAAGUG-----------GAGA-AGAGGGCCAAGAUUAACGGAAGAACUGGUUCAUGAAU ...(((((((.......((((((.(((.(((...((.(((((((.((..((....))...))-----------.)))-)))).))...))).))))))))).)))))))...... ( -34.90) >DroPse_CAF1 130199 101 + 1 GGCGAACCAGAAGGUUAUCUUCUUGUUGGUUGUCGUGUUCUUCUGCAGAGAGG-AGAGAGCC-----------GAGA-GAAAGG-GAAUAUUAACGGAAGAACUGUCUUAUGAAU ((((((((....)))).((((((.((((((..((...(((((((....)))))-))....((-----------....-....))-))..))))))))))))..))))........ ( -23.60) >DroEre_CAF1 117793 110 + 1 GGCGAACCAGAAGGUUAUCUUCUUGUUCGUUGUCGUGUUCUUCUGCAGAUCGAAAGAAAGAAGA----CGGAAGAGA-AGAGGGGUCAGAUUAACGGAAGAACUGGUUCAUGAAU ...(((((((..(..(.(((((((.((((((.((.(((......)))..((....))..)).))----)))).))))-)))..)..).......(....)..)))))))...... ( -34.60) >DroWil_CAF1 165675 99 + 1 GGCAAACCAGAAGGUUAUCUUCUUAUUGGUUGUAGUGUUCUUCUGCAAA---AAAGAAAGAG------------AGA-UGGAGAGAACGAUUAACGGAAGAACUGGCUCAUGAAU (((.((((....)))).((((((.....((((...((((((.((.((..---..........------------...-)).))))))))..))))))))))....)))....... ( -19.36) >DroYak_CAF1 115607 103 + 1 GGCGAACCAGAAGGUUAUCUUCUUGUUUGUUGUCGUGUUCUUCUGCAGAUCGAAAGAAAGAG-----------GAGA-GGAGGGGCGAGAUUAACGGAAGAACUGGUUUAUGAAU ...(((((((..........((((((((.((.((....((((((.....((....)).))))-----------))))-.)).))))))))....(....)..)))))))...... ( -29.30) >DroAna_CAF1 118035 115 + 1 GGCGAACCAGAAGGUUAUCUUCUUGUUCGUUGUCGUGUUCUUCUGCAGGUCCAAAAAAGAGAGAGAAAAAUAAGAGAAAAAGGGUCGAGAUUAACGGAAGAACUGGCUUAUGAAU (((((((.(((((.....))))).)))))))((((.(((((((((..(..((.............................))..)........)))))))))))))........ ( -29.25) >consensus GGCGAACCAGAAGGUUAUCUUCUUGUUCGUUGUCGUGUUCUUCUGCAGAUCGAAAGAAAGAG___________GAGA_AGAGGGGCAAGAUUAACGGAAGAACUGGCUCAUGAAU (((((((.(((((.....))))).))))))).....(((((((((..((((.....................................))))..)))))))))............ (-20.67 = -20.97 + 0.31)

| Location | 20,860,672 – 20,860,775 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -14.00 |
| Energy contribution | -13.91 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
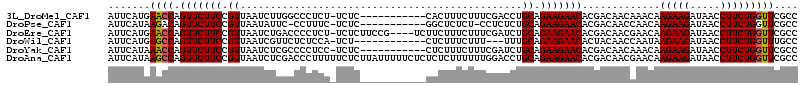
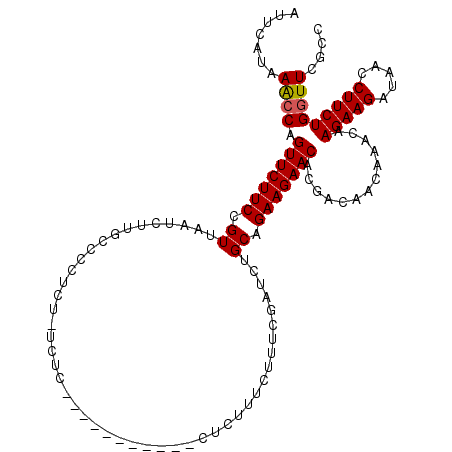
>3L_DroMel_CAF1 20860672 103 - 23771897 AUUCAUGAACCAGUUCUUCCGUUAAUCUUGGCCCUCU-UCUC-----------CACUUUCUUUCGACCUGCAGAAGAACACGACAACAAACAAGAAGAUAACCUUCUGGUUCGCC ......(((((((.(((((.(((....(((....(((-(((.-----------((...((....))..)).))))))...))).....)))..))))).......)))))))... ( -20.20) >DroPse_CAF1 130199 101 - 1 AUUCAUAAGACAGUUCUUCCGUUAAUAUUC-CCUUUC-UCUC-----------GGCUCUCU-CCUCUCUGCAGAAGAACACGACAACCAACAAGAAGAUAACCUUCUGGUUCGCC ............(((((((.((........-((....-....-----------))......-.......)).))))))).(((..((((....((((.....))))))))))).. ( -14.90) >DroEre_CAF1 117793 110 - 1 AUUCAUGAACCAGUUCUUCCGUUAAUCUGACCCCUCU-UCUCUUCCG----UCUUCUUUCUUUCGAUCUGCAGAAGAACACGACAACGAACAAGAAGAUAACCUUCUGGUUCGCC ......(((((.(((((((.((..(((..........-.........----.............)))..)).))))))).............(((((.....))))))))))... ( -22.05) >DroWil_CAF1 165675 99 - 1 AUUCAUGAGCCAGUUCUUCCGUUAAUCGUUCUCUCCA-UCU------------CUCUUUCUUU---UUUGCAGAAGAACACUACAACCAAUAAGAAGAUAACCUUCUGGUUUGCC ......(((((.(((((((.((...............-...------------..........---...)).))))))).............(((((.....))))))))))... ( -17.08) >DroYak_CAF1 115607 103 - 1 AUUCAUAAACCAGUUCUUCCGUUAAUCUCGCCCCUCC-UCUC-----------CUCUUUCUUUCGAUCUGCAGAAGAACACGACAACAAACAAGAAGAUAACCUUCUGGUUCGCC .......((((.(((((((.((..(((..........-....-----------...........)))..)).))))))).............(((((.....))))))))).... ( -18.11) >DroAna_CAF1 118035 115 - 1 AUUCAUAAGCCAGUUCUUCCGUUAAUCUCGACCCUUUUUCUCUUAUUUUUCUCUCUCUUUUUUGGACCUGCAGAAGAACACGACAACGAACAAGAAGAUAACCUUCUGGUUCGCC ............(((((((.((...((.(((..............................)))))...)).))))))).......(((((.(((((.....))))).))))).. ( -20.41) >consensus AUUCAUAAACCAGUUCUUCCGUUAAUCUUGCCCCUCU_UCUC___________CUCUUUCUUUCGAUCUGCAGAAGAACACGACAACAAACAAGAAGAUAACCUUCUGGUUCGCC .......((((.(((((((.((...............................................)).))))))).............(((((.....))))))))).... (-14.00 = -13.91 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:15 2006