
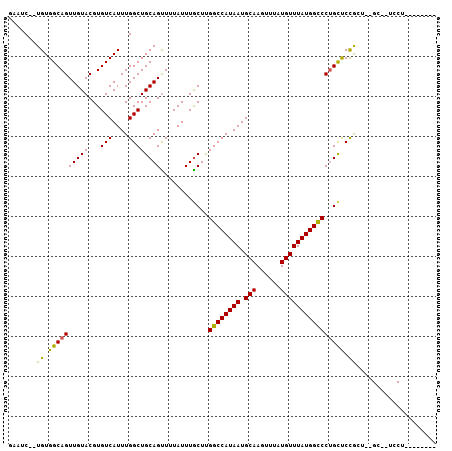
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,812,649 – 20,812,749 |
| Length | 100 |
| Max. P | 0.701037 |

| Location | 20,812,649 – 20,812,749 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20812649 100 + 23771897 GAAUC--UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUGCUUGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCAGCUCCGAAGGGC--UCCUCGCAU--- (((.(--((..((...((.......))....))..))).)))....(((.((((((((.(((......))))))))))).))).((....)).--.........--- ( -29.10) >DroPse_CAF1 80559 87 + 1 CAAUC--UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUGCUUGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCUGCUCUGCU------------------ ....(--((..((...((.......))....))..)))........((..((((((((.(((......)))))))))))..))......------------------ ( -24.60) >DroEre_CAF1 69382 103 + 1 GAAUC--UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUGCUCGGCCAUAAUGCAAGUUUAUGUUUAUGGUCCUGCUCCGAAGGGC--UCCUCGCUCCUC (((.(--((..((...((.......))....))..))).)))....((..((((((((.(((......)))))))))))..))...((.((((--.....)))).)) ( -26.90) >DroWil_CAF1 91796 93 + 1 ACAUCUGUGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUACUUGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCUGCUCCA----GC--UCCA-------- .........(((.(((((.....(((....))).((((............((((((((.(((......)))))))))))))))..))----))--))))-------- ( -24.10) >DroAna_CAF1 69737 102 + 1 GAUUC--UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGCUUUAUUUGCUCGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCUGUCCCACUUGGUCCUGCUCCUGC--- ((((.--.((((.(((((((...(((....))))))))))......((..((((((((.(((......)))))))))))..)).))))..))))..........--- ( -28.50) >DroPer_CAF1 81136 87 + 1 CAAUC--UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUGCUUGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCUGCUCUGCU------------------ ....(--((..((...((.......))....))..)))........((..((((((((.(((......)))))))))))..))......------------------ ( -24.60) >consensus GAAUC__UGUGGCAGUUGUACGUGUCAUUUGGCUGCAGUUUUAUUUGCUUGGCCAUAAUGCAAGUUUAUGUUUAUGGCCCUGCUCCGCU__GC__UCCU________ .......((.((((((((((...(((....))))))))............((((((((.(((......)))))))))))))))).)).................... (-20.79 = -20.10 + -0.69)


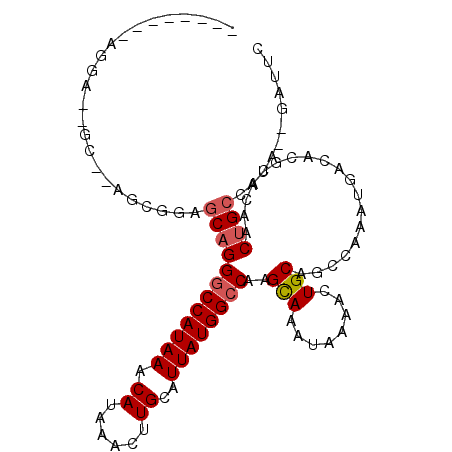
| Location | 20,812,649 – 20,812,749 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -16.77 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20812649 100 - 23771897 ---AUGCGAGGA--GCCCUUCGGAGCUGGGCCAUAAACAUAAACUUGCAUUAUGGCCAAGCAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACA--GAUUC ---.(.(((((.--...))))).)(((.((((((((.((......))..)))))))).)))..........((((....(((...)))....))))....--..... ( -24.10) >DroPse_CAF1 80559 87 - 1 ------------------AGCAGAGCAGGGCCAUAAACAUAAACUUGCAUUAUGGCCAAGCAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACA--GAUUG ------------------.((((.((..((((((((.((......))..))))))))..)).......((((((......)).)).))....))))....--..... ( -21.40) >DroEre_CAF1 69382 103 - 1 GAGGAGCGAGGA--GCCCUUCGGAGCAGGACCAUAAACAUAAACUUGCAUUAUGGCCGAGCAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACA--GAUUC ((((.((.....--)).))))(..(((((........)....((.((((((.((((...(((........))).)))))))).)).))....))))..).--..... ( -22.20) >DroWil_CAF1 91796 93 - 1 --------UGGA--GC----UGGAGCAGGGCCAUAAACAUAAACUUGCAUUAUGGCCAAGUAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACACAGAUGU --------....--.(----((..((((((((((((.((......))..))))))))...........((((((......)).)).))....))))....))).... ( -20.90) >DroAna_CAF1 69737 102 - 1 ---GCAGGAGCAGGACCAAGUGGGACAGGGCCAUAAACAUAAACUUGCAUUAUGGCCGAGCAAAUAAAGCUGCAGCCAAAUGACACGUACAACUGCCACA--GAAUC ---...(..((((......(((......((((((((.((......))..))))))))..(((........)))..........)))......))))..).--..... ( -25.50) >DroPer_CAF1 81136 87 - 1 ------------------AGCAGAGCAGGGCCAUAAACAUAAACUUGCAUUAUGGCCAAGCAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACA--GAUUG ------------------.((((.((..((((((((.((......))..))))))))..)).......((((((......)).)).))....))))....--..... ( -21.40) >consensus ________AGGA__GC__AGCGGAGCAGGGCCAUAAACAUAAACUUGCAUUAUGGCCAAGCAAAUAAAACUGCAGCCAAAUGACACGUACAACUGCCACA__GAUUC ........................((((((((((((.((......))..))))))))..(((........)))...................))))........... (-16.77 = -17.13 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:04 2006