| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,334,814 – 2,334,911 |
| Length | 97 |
| Max. P | 0.626105 |

| Location | 2,334,814 – 2,334,911 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2334814 97 + 23771897 AAC-UAUGGCGUCUGUUAGAGCAAAGUU----AGUUUUGCAGC-------GGGAUAUUCUUCAAACUUUUCA---CUCACUUCCGCCGCUGGCAAAAAAAAGGCGUCAAGUA .((-(.(((((((((((((.((((((..----..)))))).((-------((((..................---.....))))))..))))).......))))))))))). ( -24.61) >DroSec_CAF1 84060 95 + 1 AAC-UAUGGCGUCUGUUA--GCAAAGUU----AGUUUUGCAGC-------GGAAUAUUCUUCAAACUUUUCA---CUCACUUCCGCCGCUGGCAAAAAAAAGGCGUCAAGUA .((-(.((((((((((((--((...((.----......)).((-------((((..................---.....)))))).)))))).......))))))))))). ( -23.91) >DroSim_CAF1 84233 95 + 1 AAC-UAUGGCGUCUGUUA--GCAAAGUU----AGUUUUGCAGC-------GGAAUAUUCUUCAAACUUUUCA---CUCAGUUCCGCCGCCGGCAAAAAAAAGGCGUCAAGUA .((-(.((((((((....--((((((..----..)))))).((-------(((((.................---....)))))))..............))))))))))). ( -25.00) >DroEre_CAF1 89329 98 + 1 AAC-UAUGGCGUCUGUUA--GCAAAGUUAGUUAGUUUUGCAGC-------GGAAUAUUCUUCAAACUUUUUA---CUCACUUCCGCCGCUGGCAAAA-CAAGGCGUCAAGUA .((-(.((((((((..((--((.......))))((((((((((-------((....................---..........))))).))))))-).))))))))))). ( -28.31) >DroYak_CAF1 86955 94 + 1 AAC-UAUGGCGUCUGUUA--GCAAAGUU----AGUUUUGCAGC-------GGAAUAUUCUUCAAACUUUUCA---CUCACUUCCGCCGCUGGCAAAA-CAAGGCGUCAAGUA .((-(.((((((((..((--((...)))----)((((((((((-------((....................---..........))))).))))))-).))))))))))). ( -27.01) >DroAna_CAF1 80762 94 + 1 ACUAAAUGGUGUCGGCUG--GGAAAGUU----UACUUUGUAACAGCGACAGGAGAAUUCUUGGAACUUUUUGCAGCUCACUCCAG------------GAAAGGCAACGAGUA (((...(((.((.(((((--((((((((----(...((((....))))((((......))))))))))))).))))).)).))).------------....(....).))). ( -25.60) >consensus AAC_UAUGGCGUCUGUUA__GCAAAGUU____AGUUUUGCAGC_______GGAAUAUUCUUCAAACUUUUCA___CUCACUUCCGCCGCUGGCAAAAAAAAGGCGUCAAGUA ......((((((((......((((((........))))))............................................(((...))).......)))))))).... (-14.84 = -14.90 + 0.06)

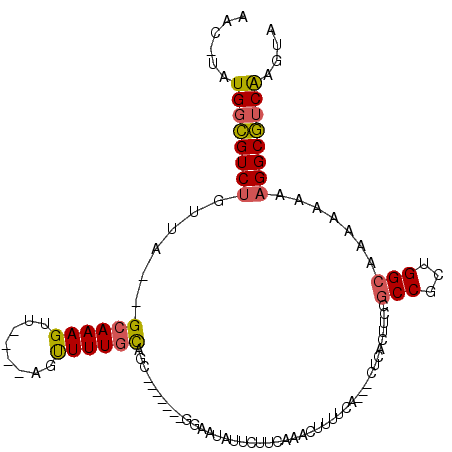

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:58 2006