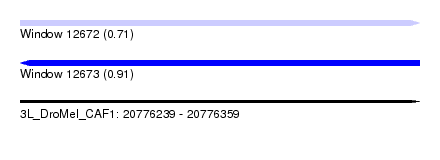
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,776,239 – 20,776,359 |
| Length | 120 |
| Max. P | 0.914960 |

| Location | 20,776,239 – 20,776,359 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -26.16 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
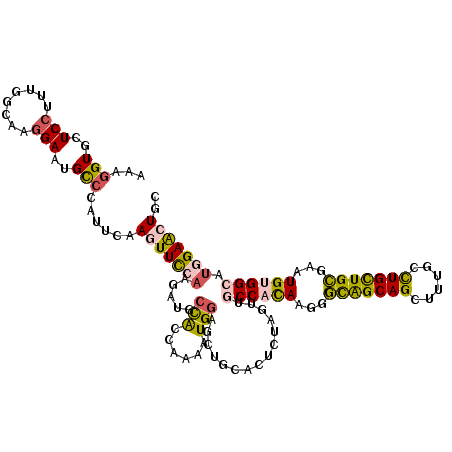
>3L_DroMel_CAF1 20776239 120 + 23771897 AAAGGUGCUCCUUUGGAAAGGAAUGCCCCUUCAAGUUCCAAGAUGCUACCAAAAUGGAGCUCCACUGUAGUUGCCAUAAGGGUGGCAGUUUUUCCUGCUGCGAAUGUGGCAUGGAACUGC .((((.(((((((....)))))..)).))))..(((((((....(((((.....(((....)))..)))))(((((((...(..((((......))))..)...)))))))))))))).. ( -42.60) >DroPse_CAF1 37786 120 + 1 UUAGGUGUUCCGCGAGCAAGGAGUGCCUAUUUAAGUUCGAGGAUGCCAGCAAGAUGGAGGUGCAUCGUAGAUGCCACAAGGGUAGCAGUUUCGCCUGCUGCGAAUGCGGCUUGGAGAUGG .((((..((((........))))..))))......(((((.((((((..(......)..).)))))......(((.((...(((((((......)))))))...)).))))))))..... ( -38.60) >DroSim_CAF1 30633 120 + 1 AAAGGUGCUCCUUUGGAAAGGAAUGCCCCUUCAAGUUCCAAGAUGCUACCAGAAUGGAGCUUCACUCCAGUUGCCAUAAGGGUGGCAGCUUUUCCUGCUGCGAAUGUGGCAUGGAACUGC .((((.(((((((....)))))..)).))))..(((((((...(((((((((..(((((.....)))))((((((((....))))))))........))).....))))))))))))).. ( -44.00) >DroYak_CAF1 30105 120 + 1 AAAGGUGCUCCUUUGGGAAGGAAUGUCCCUUCAAGUUUCAAGAUGCUGCCAAAAUGAAGCUCCACUCCAGUUGCCACAAGGGCGGCAGCUUUUCCUGUUGCGAAUGUGGAAUGGAACUGC ...((.((((.((((((((((......))))).(((........))).)))))..).))).))..((((....(((((...(((((((......)))))))...)))))..))))..... ( -38.50) >DroAna_CAF1 33305 120 + 1 UUAGGUGCUCCUUUGGCAAAGACUGUACAUUCAAGUUCCUGGAUGCCAAGAAAAUGGAACUGCACUCGAGGUGCCACAAGGACAGCAGCUUUGCUUGCUGUGAGUGUGGACUGGAGCUCC ...((.(((((.........((.((((..((((..(((.(((...))).)))..))))..)))).))..(((.(((((...((((((((...)).))))))...))))))))))))).)) ( -40.70) >DroPer_CAF1 37502 120 + 1 UUAGGUGUUCCGCGAGCAAGGAGUGCCUAUUUAAGUUCGAGGAUGCCAGCAAGAUGGAGGUGCAUCGUAGAUGCCACAAGGGCAGCAGUUUCGCCUGCUGCGAAUGCGGCUUGGAGAUGG .((((..((((........))))..))))......(((((.((((((..(......)..).)))))......(((.((...(((((((......)))))))...)).))))))))..... ( -40.60) >consensus AAAGGUGCUCCUUUGGCAAGGAAUGCCCAUUCAAGUUCCAAGAUGCCACCAAAAUGGAGCUGCACUCUAGUUGCCACAAGGGCAGCAGCUUUGCCUGCUGCGAAUGUGGCAUGGAACUGC ...(((..(((........)))..)))......(((((((.....(((......)))...............((((((...(((((((......)))))))...)))))).))))))).. (-26.16 = -25.88 + -0.27)

| Location | 20,776,239 – 20,776,359 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
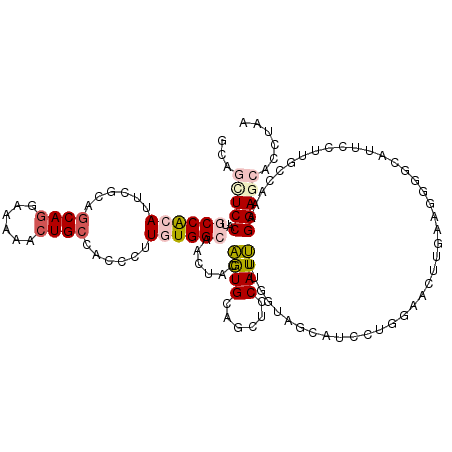
>3L_DroMel_CAF1 20776239 120 - 23771897 GCAGUUCCAUGCCACAUUCGCAGCAGGAAAAACUGCCACCCUUAUGGCAACUACAGUGGAGCUCCAUUUUGGUAGCAUCUUGGAACUUGAAGGGGCAUUCCUUUCCAAAGGAGCACCUUU ..((((((((((((........((((......))))........)))))((((.(((((....))))).)))).......))))))).(((((.((..(((((....))))))).))))) ( -39.39) >DroPse_CAF1 37786 120 - 1 CCAUCUCCAAGCCGCAUUCGCAGCAGGCGAAACUGCUACCCUUGUGGCAUCUACGAUGCACCUCCAUCUUGCUGGCAUCCUCGAACUUAAAUAGGCACUCCUUGCUCGCGGAACACCUAA .....(((..((((((...(.(((((......))))).)...))))))......((((......))))..((.((((........((.....))........)))).)))))........ ( -30.69) >DroSim_CAF1 30633 120 - 1 GCAGUUCCAUGCCACAUUCGCAGCAGGAAAAGCUGCCACCCUUAUGGCAACUGGAGUGAAGCUCCAUUCUGGUAGCAUCUUGGAACUUGAAGGGGCAUUCCUUUCCAAAGGAGCACCUUU ..((((((((((((.....(((((.......)))))........)))))((..(((((......)))))..)).......))))))).(((((.((..(((((....))))))).))))) ( -41.62) >DroYak_CAF1 30105 120 - 1 GCAGUUCCAUUCCACAUUCGCAACAGGAAAAGCUGCCGCCCUUGUGGCAACUGGAGUGGAGCUUCAUUUUGGCAGCAUCUUGAAACUUGAAGGGACAUUCCUUCCCAAAGGAGCACCUUU (((((((((((((...(((......)))..((.((((((....)))))).)))))))))))))((.(((((((((....)))......((((((....))))))))))))))))...... ( -39.10) >DroAna_CAF1 33305 120 - 1 GGAGCUCCAGUCCACACUCACAGCAAGCAAAGCUGCUGUCCUUGUGGCACCUCGAGUGCAGUUCCAUUUUCUUGGCAUCCAGGAACUUGAAUGUACAGUCUUUGCCAAAGGAGCACCUAA ((.(((((.(((((((...((((((.((...))))))))...))))).)).....(((((....((..(((((((...)))))))..))..))))).............))))).))... ( -42.10) >DroPer_CAF1 37502 120 - 1 CCAUCUCCAAGCCGCAUUCGCAGCAGGCGAAACUGCUGCCCUUGUGGCAUCUACGAUGCACCUCCAUCUUGCUGGCAUCCUCGAACUUAAAUAGGCACUCCUUGCUCGCGGAACACCUAA .....(((..((((((...(((((((......)))))))...))))))......((((......))))..((.((((........((.....))........)))).)))))........ ( -37.29) >consensus GCAGCUCCAUGCCACAUUCGCAGCAGGAAAAACUGCCACCCUUGUGGCAACUACAGUGCAGCUCCAUUUUGGUAGCAUCCUGGAACUUGAAGGGGCAUUCCUUGCCAAAGGAGCACCUAA ...(((((..((((((......((((......))))......))))))......((((......)))).........................................)))))...... (-18.29 = -18.98 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:53 2006