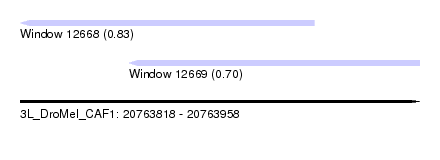
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,763,818 – 20,763,958 |
| Length | 140 |
| Max. P | 0.830989 |

| Location | 20,763,818 – 20,763,921 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.44 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -7.44 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20763818 103 - 23771897 AUUUAAUCCCAUACUCAAUAAAGUACACAAA-------------AAGCUUUCUGCUUUAACAAACG--GCUCUUAGCCGAAGAAGAUACCUUC--GAGUGGGGAAAAAGAAGUCGGAUUA ......((((.(((((..............(-------------((((.....)))))......((--((.....))))..((((....))))--)))))))))................ ( -26.80) >DroSec_CAF1 19325 116 - 1 CUUUAAUCCCAUACUCAAUAAAGUACAUAAAGCGUAGAAAAAUACAGCUUUCUGCUUUAACAAACA--GCUCUUAGCCGAAGAAGAUACCUUC--GAGUGGGAAAAAAGAAGUCGGAUUA ((((..((((((......(((((((...((((((((......))).))))).))))))).......--((.....))(((((.......))))--).))))))..))))........... ( -25.80) >DroSim_CAF1 17729 116 - 1 CUUUAAUCCCAUACUCAAUAAAGAACAUACAGCGUAGAAAAAUACAGUUUUCUGCUUUAAAAAACA--GCUCUUAGCCGAAGAAGAUAACUUC--GAGUGGAAGAAAAGAAGUCGGAUUA ...((((((...((((.................((((((((......))))))))...........--..((((.(((((((.......))))--).))..))))...).))).)))))) ( -21.50) >DroEre_CAF1 19235 117 - 1 CUGUGAUCCCUUACUCAAUUGAUUACACAAG-GGUUUGCAAAUACAGCUUUCUGCCUUUAGAAGCA--GCUCUUAGCUGGGGAAGAUACCUUCAAUAGUGGAAAAAAAGAAGGCGGAUUA .((..(.(((((................)))-)).)..))..........(((((((((.....((--((.....))))..((((....))))...............)))))))))... ( -29.59) >DroYak_CAF1 17657 85 - 1 CUUUGAUCCCUUUCUCUAUGAAUUAC---------------------------------A--CACAUUAUUCUUUGCUGAAGAAAAUACCUUCAUGAGUGGGAAAAAAGAAGUCCGAUUA ((((..((((...(((...((((...---------------------------------.--......)))).....(((((.......))))).))).))))..))))........... ( -15.30) >consensus CUUUAAUCCCAUACUCAAUAAAGUACACAAA__GU____AAAUACAGCUUUCUGCUUUAACAAACA__GCUCUUAGCCGAAGAAGAUACCUUC__GAGUGGGAAAAAAGAAGUCGGAUUA ((((..((((..((((..............................((.....)).......................((((.......))))..))))))))..))))........... ( -7.44 = -8.14 + 0.70)


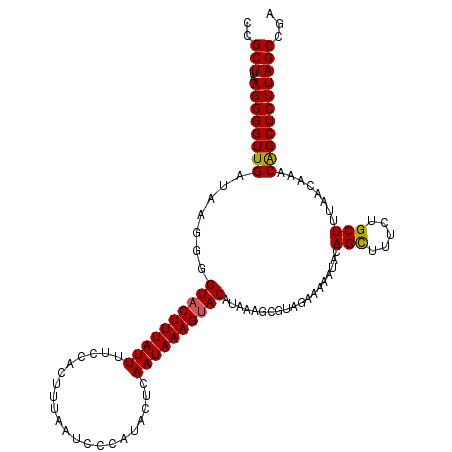
| Location | 20,763,856 – 20,763,958 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20763856 102 - 23771897 UCGCUUAAGGGGUUGAUAAGCAGUACUUUAUUUUCUAAUUUAAUCCCAUACUCAAUAAAGUACACAAA-------------AAGCUUUCUGCUUUAACAAACGGCUCUUAGCCGA ..(((..((((((((.......((((((((((.....................))))))))))....(-------------((((.....)))))......)))))))))))... ( -22.30) >DroSec_CAF1 19363 115 - 1 CCGCUUAAGGGGUUGAUAAGGGGUACUUUAUUUUCCACUUUAAUCCCAUACUCAAUAAAGUACAUAAAGCGUAGAAAAAUACAGCUUUCUGCUUUAACAAACAGCUCUUAGCCGA ..(((..((((((((....(((((..................)))))........(((((((...((((((((......))).))))).))))))).....)))))))))))... ( -26.37) >DroSim_CAF1 17767 115 - 1 CCGCUUAAGGGGUUGAUAAGGGGUACUUUAUUUUUCACUUUAAUCCCAUACUCAAUAAAGAACAUACAGCGUAGAAAAAUACAGUUUUCUGCUUUAAAAAACAGCUCUUAGCCGA ..(((..((((((((....(((((..................))))).......................((((((((......)))))))).........)))))))))))... ( -23.17) >consensus CCGCUUAAGGGGUUGAUAAGGGGUACUUUAUUUUCCACUUUAAUCCCAUACUCAAUAAAGUACAUAAAGCGUAGAAAAAUACAGCUUUCUGCUUUAACAAACAGCUCUUAGCCGA ..(((..((((((((.......((((((((((.....................))))))))))...................(((.....)))........)))))))))))... (-18.54 = -18.43 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:48 2006