| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,746,398 – 20,746,515 |
| Length | 117 |
| Max. P | 0.742995 |

| Location | 20,746,398 – 20,746,515 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
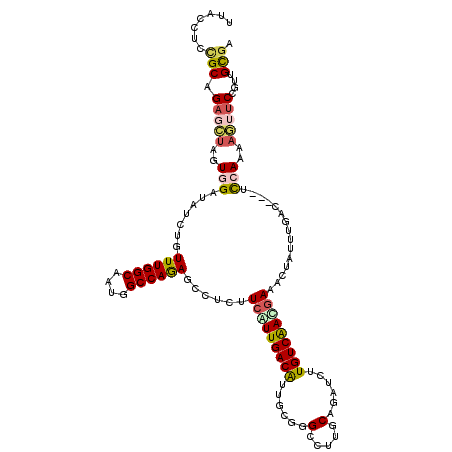
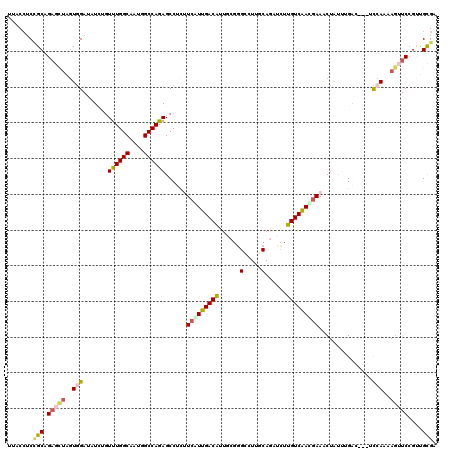
>3L_DroMel_CAF1 20746398 117 - 23771897 UUACCUCCGCAGAGCUAGUGGAUAUCUGUUUGGCUUUGGCCAGAGCCAUUUCGUUGACAUUUCUGGCCUUGCAGAUCUUGUCAACGAAACUAUUCGAU---UCCAAAAGUUCCGUUGCGA .......(((((((((..((((.(((....(((((((....)))))))(((((((((((..((((......))))...)))))))))))......)))---))))..)))))...)))). ( -44.80) >DroVir_CAF1 5004 120 - 1 UUACAUCGGCUGUGGUGCUGGAAAUAAGUUUGGCAACCGCCAGAGCCUCAUCGUUGACAGUGCGCGACUUGCACAGCUUGUCGAUGAAGCUGCUGGGUGAAUCGACCACAUCGGAUGCGU ...((((.(.((((((((..((......))..))...((((..(((..(.((((((((((((((.....)))))....))))))))).)..))).)))).....)))))).).))))... ( -43.90) >DroSim_CAF1 2770 117 - 1 UUACCUCCGCAGAGCUAGUGGAUAUCUGUUUGGCUAUGGCCAGAGCCUCUUCGUUGACAUUACGGGCCUUGCAGAUCUUGUCAACGAAACUAUUCGAU---UCCAAAAGUUCCGUUGCGA .......(((((((((..((((.(((..((((((....)))))).....((((((((((......((...))......)))))))))).......)))---))))..)))))...)))). ( -38.50) >DroEre_CAF1 5279 117 - 1 UUACCUCCGCUGAGCUAGUGGAUAUCUUUUUGGCAAUAGCCAGAGCCUCUUCAUUGACGUUGCGGGCCUUGCAGAUCUUGUCAACGAAACUAUGUGAC---CCCAAAAGUUCCGUUGCGA .......(((.(((((..(((......(((((((....)))))))....(((.(((((((((((.....)))))....)))))).)))..........---.)))..)))))....))). ( -29.20) >DroWil_CAF1 5361 117 - 1 UUACCUCUGCCGAAGAUGUUGAUAUAAGUUUGGCAAUGGCCAAAGCCUCAUCAUUGACAUUUCGGGAUUUACAGGAUUUGUCAAUAAAACUAUUUGGC---GUUAAUAGAUCCGAUGUGC .(((.((..((((.(((((..((((.((((((((....))))))..)).)).))..)))))))))........(((((((((((.........)))))---)......))))))).))). ( -28.50) >DroYak_CAF1 2780 117 - 1 AUACCUCCGCAGAGCUAGUGGAUAUCUGUUUGGCAAUGGCCAGAGCCUCUUCAUUGACGUUGCGGGUCUUGCAGAUCUUGUCAACGAAACUAUGUGGC---CCCAAAAGUUCCGUUGCGA .......(((((((((..(((.......((((((....))))))(((....(((...(((((((((((.....)))).)).))))).....))).)))---.)))..)))))...)))). ( -34.90) >consensus UUACCUCCGCAGAGCUAGUGGAUAUCUGUUUGGCAAUGGCCAGAGCCUCUUCAUUGACAUUGCGGGCCUUGCAGAUCUUGUCAACGAAACUAUUUGAC___UCCAAAAGUUCCGUUGCGA .......(((.(((((..(((.......((((((....))))))......(((((((((......(.....)......)))))))))...............)))..)))))....))). (-21.90 = -22.43 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:44 2006