
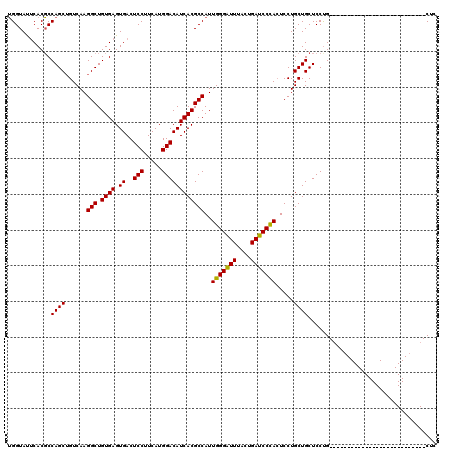
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,735,018 – 20,735,191 |
| Length | 173 |
| Max. P | 0.994888 |

| Location | 20,735,018 – 20,735,111 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -25.75 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20735018 93 + 23771897 UGGUAUUCACGCCAGCUGUCAAGGCUGUGAGUGACUCCUUCAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCAAUCCUGCUGCUCCCG---------------------------CUC .(((..(((((((.........))).))))((((.(((.....)))..)))))))(((((((((....)))))))))......((....)---------------------------).. ( -29.00) >DroSec_CAF1 10482 93 + 1 UGGUAUUCACGCCAGCUGUCAAGGCUGUGAGUGACUCCUUCAUGGACAUCACGCCAUUGGGGUUUACUGAUCCCACUCCUGCUGCUCCUG---------------------------CUC ((((..(((((((.........))).))))((((.(((.....)))..)))))))).(((((((....)))))))........((....)---------------------------).. ( -27.80) >DroSim_CAF1 13612 93 + 1 UGGUAUUCACGCCAGCUGUCAAGGCUGUGAGUGACUCCUUCAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCACUCCUGCUGCUCCUG---------------------------CUC ((((..(((((((.........))).))))((((.(((.....)))..)))))))).(((((((....)))))))........((....)---------------------------).. ( -28.40) >DroYak_CAF1 12267 120 + 1 GGGUACACACCCCAGCGGUCAAGGCUGUGAGUGACUCCUUCAUGGACAUCACGCCAUUGGGAUUUACUGAUCCUACUCCUGCUGCUCCUGCUCAUGCUGCUCCCGCUCCGGUUUCCGCUC ((((....)))).(((((...((((((.(((((..(((.....)))......((...(((((((....))))))).....((.((....))....)).))...)))))))))))))))). ( -35.90) >consensus UGGUAUUCACGCCAGCUGUCAAGGCUGUGAGUGACUCCUUCAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCACUCCUGCUGCUCCUG___________________________CUC ............((((......(((.((((.((..(((.....))))))))))))..(((((((....))))))).....)))).................................... (-25.75 = -25.38 + -0.38)


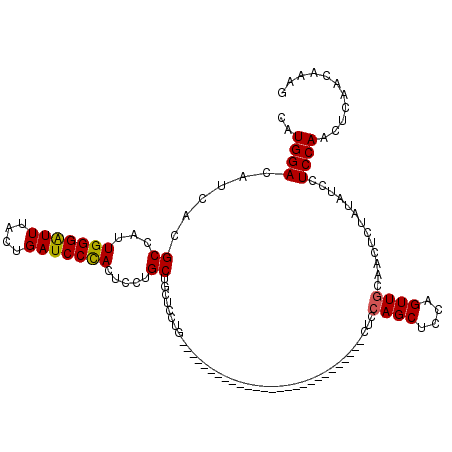
| Location | 20,735,058 – 20,735,151 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20735058 93 + 23771897 CAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCAAUCCUGCUGCUCCCG---------------------------CUCCAGCUCCAGUUGCAACUCUAUAUCCUCCAACUCAACAAAG ..((((.........(((((((((....)))))))))..(((.(((...(---------------------------(....))...))).)))...........))))........... ( -20.70) >DroSec_CAF1 10522 93 + 1 CAUGGACAUCACGCCAUUGGGGUUUACUGAUCCCACUCCUGCUGCUCCUG---------------------------CUCCAGCUCCAGUUGAAACUGUAUAUCCUCCAACUCAACAAAG ..((((...........(((((((....)))))))...(((..((....)---------------------------)..))).))))(((((..................))))).... ( -19.47) >DroSim_CAF1 13652 93 + 1 CAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCACUCCUGCUGCUCCUG---------------------------CUCCAGCUCCAGUUGUAACUCUAUAUCCUCCAACUCAACAAAG ..((((............((((((....))))))((..(((..(((....---------------------------....)))..)))..))............))))........... ( -18.70) >DroYak_CAF1 12307 120 + 1 CAUGGACAUCACGCCAUUGGGAUUUACUGAUCCUACUCCUGCUGCUCCUGCUCAUGCUGCUCCCGCUCCGGUUUCCGCUCCAGCUCCUGUUACAACUCUGUAUCCUCCAGCUCAACAAAG ..((((......(((..(((((((....))))))).....((.((....((....)).))....))...)))......))))(((.....((((....))))......)))......... ( -21.00) >consensus CAUGGACAUCACGCCAUUGGGAUUUACUGAUCCCACUCCUGCUGCUCCUG___________________________CUCCAGCUCCAGUUGCAACUCUAUAUCCUCCAACUCAACAAAG ..((((......((...(((((((....))))))).....))......................................((((....)))).............))))........... (-14.38 = -14.25 + -0.12)

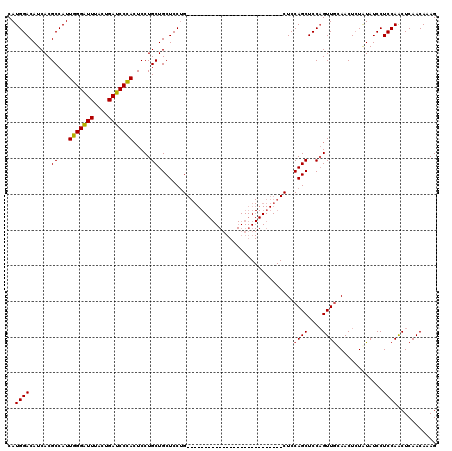
| Location | 20,735,098 – 20,735,191 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20735098 93 + 23771897 GCUGCUCCCG---------------------------CUCCAGCUCCAGUUGCAACUCUAUAUCCUCCAACUCAACAAAGUUUAAUGGAGCUCGAAGAGGACAGCAUUGUAAGGGAGAUG ....((((((---------------------------(....))....((((...((((.....((((((((......)))....))))).....))))..)))).......)))))... ( -24.40) >DroSec_CAF1 10562 93 + 1 GCUGCUCCUG---------------------------CUCCAGCUCCAGUUGAAACUGUAUAUCCUCCAACUCAACAAAGUUUAAUGGAGCUGGAAGAGGACAGCAUUGUAAGGGAGAUG ((((.((((.---------------------------.((((((((((....(((((.....................)))))..))))))))))..))))))))............... ( -34.70) >DroSim_CAF1 13692 93 + 1 GCUGCUCCUG---------------------------CUCCAGCUCCAGUUGUAACUCUAUAUCCUCCAACUCAACAAAGUUUAAUGGAGCUGGAAGAGGACAGCAUUGUGAGGGAGAUG ((((.((((.---------------------------.((((((((((.....((((.....................))))...))))))))))..))))))))............... ( -31.90) >DroYak_CAF1 12347 120 + 1 GCUGCUCCUGCUCAUGCUGCUCCCGCUCCGGUUUCCGCUCCAGCUCCUGUUACAACUCUGUAUCCUCCAGCUCAACAAAGUUCAAUGGAGCUGGAAGACGACAGCAUUGUGAGGGAGAUG ((.((....))....))..((((((((..(((....)))..))).....((((((..((((.((.((((((((..............)))))))).))..))))..)))))))))))... ( -38.74) >consensus GCUGCUCCUG___________________________CUCCAGCUCCAGUUGCAACUCUAUAUCCUCCAACUCAACAAAGUUUAAUGGAGCUGGAAGAGGACAGCAUUGUAAGGGAGAUG ....(((((.............................((((((((((.....((((.....................))))...)))))))))).....((......))..)))))... (-23.29 = -23.60 + 0.31)

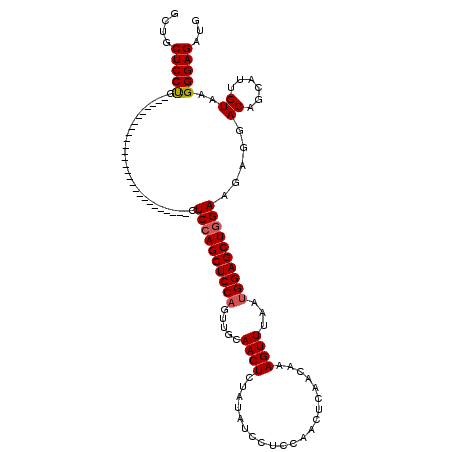
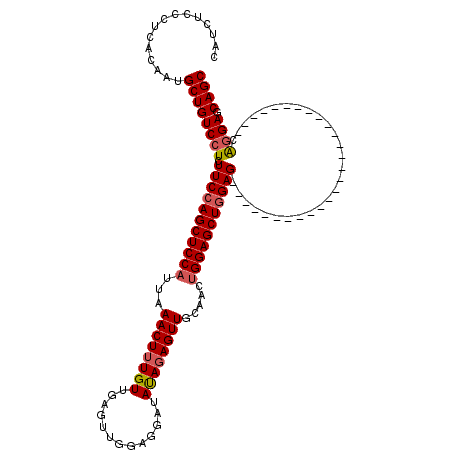
| Location | 20,735,098 – 20,735,191 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.89 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20735098 93 - 23771897 CAUCUCCCUUACAAUGCUGUCCUCUUCGAGCUCCAUUAAACUUUGUUGAGUUGGAGGAUAUAGAGUUGCAACUGGAGCUGGAG---------------------------CGGGAGCAGC ...............((((((((.(((.(((((((...((((((((.............)))))))).....))))))).)))---------------------------.)))).)))) ( -31.42) >DroSec_CAF1 10562 93 - 1 CAUCUCCCUUACAAUGCUGUCCUCUUCCAGCUCCAUUAAACUUUGUUGAGUUGGAGGAUAUACAGUUUCAACUGGAGCUGGAG---------------------------CAGGAGCAGC ...............((((((((.((((((((((......(......)((((((((.........))))))))))))))))))---------------------------.)))).)))) ( -34.30) >DroSim_CAF1 13692 93 - 1 CAUCUCCCUCACAAUGCUGUCCUCUUCCAGCUCCAUUAAACUUUGUUGAGUUGGAGGAUAUAGAGUUACAACUGGAGCUGGAG---------------------------CAGGAGCAGC ...............((((((((.(((((((((((...((((((((.............)))))))).....)))))))))))---------------------------.)))).)))) ( -34.72) >DroYak_CAF1 12347 120 - 1 CAUCUCCCUCACAAUGCUGUCGUCUUCCAGCUCCAUUGAACUUUGUUGAGCUGGAGGAUACAGAGUUGUAACAGGAGCUGGAGCGGAAACCGGAGCGGGAGCAGCAUGAGCAGGAGCAGC ...((((...(((((.((((.((((((((((((.((........)).)))))))))))))))).))))).......(((.(.(((....)....((....)).)).).))).)))).... ( -45.80) >consensus CAUCUCCCUCACAAUGCUGUCCUCUUCCAGCUCCAUUAAACUUUGUUGAGUUGGAGGAUAUAGAGUUGCAACUGGAGCUGGAG___________________________CAGGAGCAGC ...............((((((((.(((((((((((...((((((((.............)))))))).....)))))))))))............................)))).)))) (-25.27 = -25.89 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:34 2006