| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,705,208 – 20,705,356 |
| Length | 148 |
| Max. P | 0.770670 |

| Location | 20,705,208 – 20,705,321 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.59 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20705208 113 - 23771897 UGCUAGCAGCUGAUGCAUCAGCUGCUAAGAUUCCAUGCGCGCAUCUCGUGAGUAGGACAC-AAUUUAAAUGUAUCAAGUAAUUUGUAAAAAUAUCUACAGAUCUGACUGUUAAA---- (((((((((((((....))))))))))....(((.(((.(((.....))).))))))...-.........)))...(((.(((((((........)))))))...)))......---- ( -29.20) >DroSec_CAF1 1872 113 - 1 UGCUACCAGCUGAUGCGUCAGCUGCUGAGAUUCCAUGCGCGCACCUCGUGAGUCAGGCAC-AAUUUAAAUAUAUCAAGCAAUUUGUGAAAAUAUUUACAGAUCUGACUGUUAAA---- ..((..(((((((....)))))))...))........((((.....))))((((((((..-................)).(((((((((....)))))))))))))))......---- ( -26.07) >DroSim_CAF1 1859 113 - 1 UGCUAGCAGCUGAUGCAUCAGCUGCUGAGAUUCCAUGCGCGCAUCUCGUGAGUCAGGCAC-AAUUUAAAUAUAUCAAGCAAUUUGUGAAAAUAUCUACAUAUCUGACUGUUAAA---- ....(((((((((....)))))))))(((((.((....).).)))))...((((((.(((-((...................)))))...((((....))))))))))......---- ( -27.51) >DroEre_CAF1 27436 118 - 1 UGCUAGCAGCUGAUGCGUCAGCUGCUGUGAUUUCACGCGCCCAUCUCGUGAGUCACGCAUGAACUUGAAUAAAGCCAUCAAUUUAUAAGCAGAUAUUUUGUUCUGACUGUUAAAAAUA (((.(((((((((....)))))))))((((((.((((.(.....).)))))))))))))(((.(((.....)))...))).((((..((((((........)))).))..)))).... ( -33.80) >consensus UGCUAGCAGCUGAUGCAUCAGCUGCUGAGAUUCCAUGCGCGCAUCUCGUGAGUCAGGCAC_AAUUUAAAUAUAUCAAGCAAUUUGUAAAAAUAUCUACAGAUCUGACUGUUAAA____ ...((((((((((....))))))))))......((((.(.....).))))((((((((...................)).(((((((........))))))))))))).......... (-20.15 = -20.59 + 0.44)


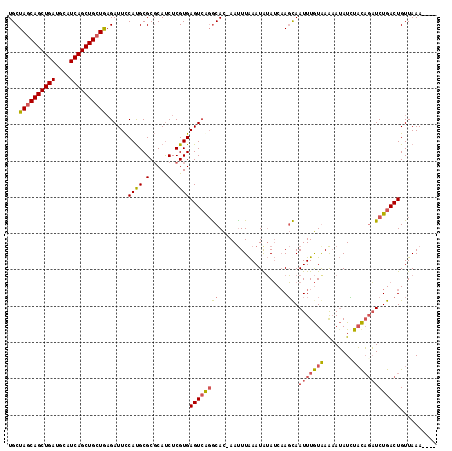
| Location | 20,705,244 – 20,705,356 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.04 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -27.80 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20705244 112 + 23771897 CUUGAUACAUUUAAAUU-GUGUCCUACUCACGAGAUGCGCGCAUGGAAUCUUAGCAGCUGAUGCAUCAGCUGCUAGCACUGAAUUUCACGACGAGUUGGCAACACAAAGCCGA ...((((((.......)-)))))..((((.....(((....)))(((((((((((((((((....)))))))))))....).))))).....))))((((........)))). ( -31.40) >DroSec_CAF1 1908 112 + 1 CUUGAUAUAUUUAAAUU-GUGCCUGACUCACGAGGUGCGCGCAUGGAAUCUCAGCAGCUGACGCAUCAGCUGGUAGCACUGAAUUUCUCGCCGAGUUGGCAGCACGAACCCGA ...............((-(((((..((((.((((((((..((.(((....))).(((((((....))))))))).)))).......))))..))))..)..))))))...... ( -34.21) >DroSim_CAF1 1895 112 + 1 CUUGAUAUAUUUAAAUU-GUGCCUGACUCACGAGAUGCGCGCAUGGAAUCUCAGCAGCUGAUGCAUCAGCUGCUAGCACUGAAUUUCUCGCAGAGUUGGCAGCACGAAACCGA ...............((-(((((..((((.(((((..((.((...(.....)(((((((((....))))))))).)).).)....)))))..))))..)..))))))...... ( -36.50) >DroEre_CAF1 27476 113 + 1 AUGGCUUUAUUCAAGUUCAUGCGUGACUCACGAGAUGGGCGCGUGAAAUCACAGCAGCUGACGCAUCAGCUGCUAGCACUGAAUUCUUCGCCGAGUUGGCAGCACCGCGCUGA .((((...(((((.((((((.((((...))))..))))))(((((....)))(((((((((....))))))))).))..))))).....))))......((((.....)))). ( -41.70) >consensus CUUGAUAUAUUUAAAUU_GUGCCUGACUCACGAGAUGCGCGCAUGGAAUCUCAGCAGCUGACGCAUCAGCUGCUAGCACUGAAUUUCUCGCCGAGUUGGCAGCACGAAGCCGA ..................(((((((((((.(((((((....)))........(((((((((....)))))))))............))))..)))))))..))))........ (-27.80 = -27.93 + 0.13)


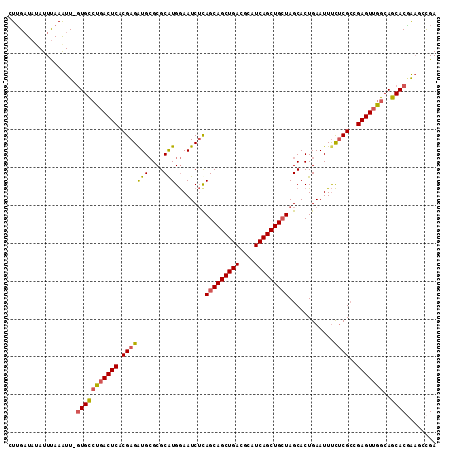
| Location | 20,705,244 – 20,705,356 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.04 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -29.55 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20705244 112 - 23771897 UCGGCUUUGUGUUGCCAACUCGUCGUGAAAUUCAGUGCUAGCAGCUGAUGCAUCAGCUGCUAAGAUUCCAUGCGCGCAUCUCGUGAGUAGGACAC-AAUUUAAAUGUAUCAAG ......((((((..((.(((((.((.((......((((((((((((((....)))))))))).(....)..))))...)).))))))).))))))-))............... ( -34.90) >DroSec_CAF1 1908 112 - 1 UCGGGUUCGUGCUGCCAACUCGGCGAGAAAUUCAGUGCUACCAGCUGAUGCGUCAGCUGCUGAGAUUCCAUGCGCGCACCUCGUGAGUCAGGCAC-AAUUUAAAUAUAUCAAG ..((((.(((((.(((.....)))(.(((.(((((......(((((((....)))))))))))).))))..))))).)))).(((.......)))-................. ( -33.80) >DroSim_CAF1 1895 112 - 1 UCGGUUUCGUGCUGCCAACUCUGCGAGAAAUUCAGUGCUAGCAGCUGAUGCAUCAGCUGCUGAGAUUCCAUGCGCGCAUCUCGUGAGUCAGGCAC-AAUUUAAAUAUAUCAAG ........(((((....((((.((((((......((((((((((((((....)))))))))).(....)..))))...))))))))))..)))))-................. ( -38.60) >DroEre_CAF1 27476 113 - 1 UCAGCGCGGUGCUGCCAACUCGGCGAAGAAUUCAGUGCUAGCAGCUGAUGCGUCAGCUGCUGUGAUUUCACGCGCCCAUCUCGUGAGUCACGCAUGAACUUGAAUAAAGCCAU .((((.....)))).......(((.....(((((((((.(((((((((....)))))))))((((((.((((.(.....).)))))))))))))).....)))))...))).. ( -40.40) >consensus UCGGCUUCGUGCUGCCAACUCGGCGAGAAAUUCAGUGCUAGCAGCUGAUGCAUCAGCUGCUGAGAUUCCAUGCGCGCAUCUCGUGAGUCAGGCAC_AAUUUAAAUAUAUCAAG ........(((((....(((((.(((((......((((((((((((((....)))))))))).(....)..))))...))))))))))..))))).................. (-29.55 = -30.18 + 0.63)

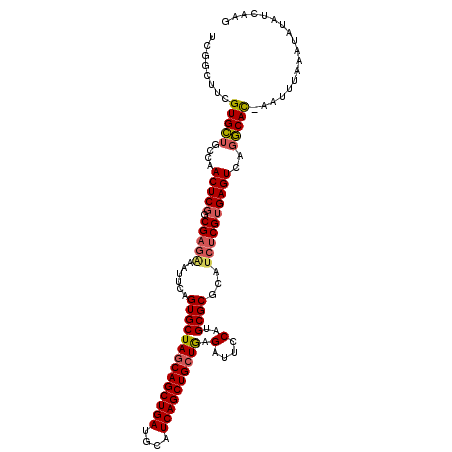

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:18 2006