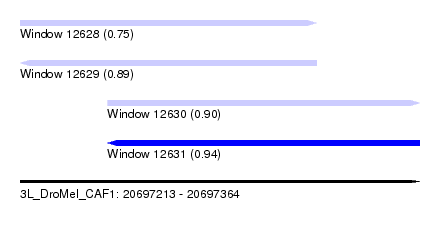
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,697,213 – 20,697,364 |
| Length | 151 |
| Max. P | 0.939985 |

| Location | 20,697,213 – 20,697,325 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -32.27 |
| Energy contribution | -32.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20697213 112 + 23771897 AUUGGCACCUGUGUGUUGCCAGUU-------GCCAGUUGCCGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUCCGCUGACCUCGUUUACGGGUCAGAGCAGC ..(((((.(.....).)))))(((-------((.((((((....))))))(((((.((((((................)))))).)-))))..((((((.((....)))))))).))))) ( -35.09) >DroEre_CAF1 19223 119 + 1 AUUGGCACCUGUGUGUUGCCAGUUGCCAGUUGCCAGUUGCCGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUCCACUGACCUCGUUUACGGGUCAGAGCAGC ..(((((.(((.(.....)))).)))))(((((.((((((....))))))(((((.((((((................)))))).)-))))..((((((.((....)))))))).))))) ( -39.29) >DroYak_CAF1 56 113 + 1 AUUGGCACCUGUGUGUUGCCAGUU-------GGCAGUUGCCGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUUUUUUCCGCUGACCUCGUUUACGGGUCAGAGCAGC ..(((((.(.....).)))))(((-------..(((((((....)))))))..)))....((((...(((.....)))...............((((((.((....)))))))).)))). ( -32.80) >consensus AUUGGCACCUGUGUGUUGCCAGUU_______GCCAGUUGCCGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU_UUUCCGCUGACCUCGUUUACGGGUCAGAGCAGC (((((((.(.....).)))))))..........(((((((....))))))).........((((...(((.....)))...............((((((.((....)))))))).)))). (-32.27 = -32.27 + 0.00)


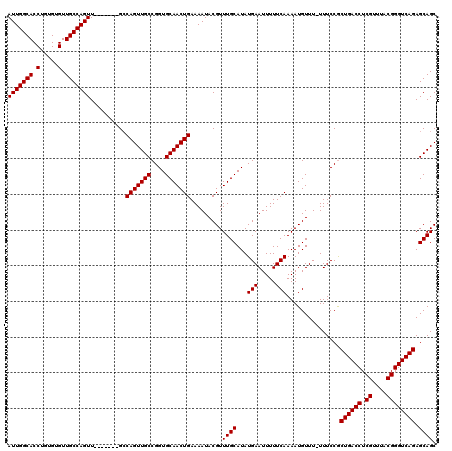
| Location | 20,697,213 – 20,697,325 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20697213 112 - 23771897 GCUGCUCUGACCCGUAAACGAGGUCAGCGGAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCGGCAACUGGC-------AACUGGCAACACACAGGUGCCAAU .(((((..((((((....)).)))))))))...-........(((((.....(((((....))))))))))(((((....)))))((((-------(.(((........))).))))).. ( -32.60) >DroEre_CAF1 19223 119 - 1 GCUGCUCUGACCCGUAAACGAGGUCAGUGGAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCGGCAACUGGCAACUGGCAACUGGCAACACACAGGUGCCAAU (((((.((((((((....)).))))))......-........(((.....)))...)))..........(((((((....)))))))))...(((((.(((........))).))))).. ( -35.90) >DroYak_CAF1 56 113 - 1 GCUGCUCUGACCCGUAAACGAGGUCAGCGGAAAAAAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCGGCAACUGCC-------AACUGGCAACACACAGGUGCCAAU .(((((..((((((....)).)))))))))...........((((((.....(((((....)))))))))))..((((((((....)))-------...((....))....))))).... ( -31.50) >consensus GCUGCUCUGACCCGUAAACGAGGUCAGCGGAAA_AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCGGCAACUGGC_______AACUGGCAACACACAGGUGCCAAU ((..(.((((((((....)).)))))).).............(((.....)))...))...........(((((((....)))))))............(((((.(.....).))))).. (-28.90 = -28.90 + 0.00)

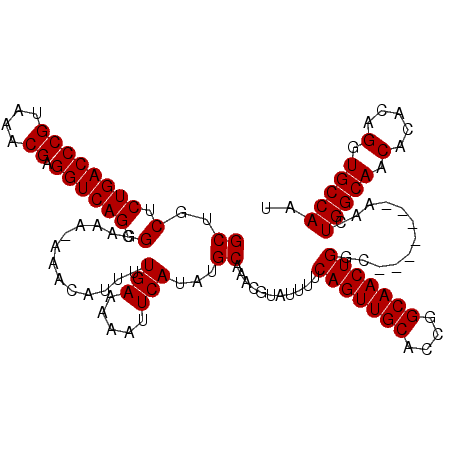
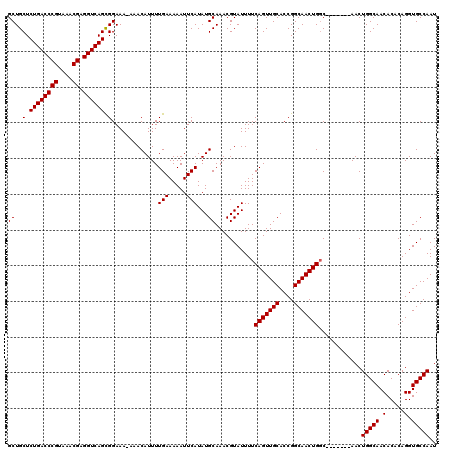
| Location | 20,697,246 – 20,697,364 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -24.72 |
| Energy contribution | -24.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20697246 118 + 23771897 CGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUCCGCUGACCUCGUUUACGGGUCAGAGCAGCUAUGAAAAGUUUGCCC-UGCAAACUAAAUUGCCGUUUGCA ...(((((.((..(((..(((((((..(((((((((((...((((.-......((((((.((....))))))))))))....)))))))))))...-)))))))...)))..)).))))) ( -32.51) >DroPse_CAF1 2 108 + 1 -----------AAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUGCGUUGACCUCGUUUACGGGUCAGAGCGGGUAUGAAAAGUUUGCCCGUACAAACUAAAUUGCCGCCUGCA -----------.....((((..((((.(((.....)))..))))..-...))))(((((.((....)))))))..((((((......((((((......))))))........)))))). ( -26.14) >DroEre_CAF1 19263 118 + 1 CGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUCCACUGACCUCGUUUACGGGUCAGAGCAGCUAUGAAAAGUUUGCCC-UGCAAACUAAAUUGCCGUUUGCA ...(((((.((..(((..(((((((..(((((((((((...((((.-......((((((.((....))))))))))))....)))))))))))...-)))))))...)))..)).))))) ( -32.51) >DroYak_CAF1 89 119 + 1 CGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUUUUUUCCGCUGACCUCGUUUACGGGUCAGAGCAGCUAUGAAAAGUUUGCCC-UGCAAACUAAAUUGCCGUUUGCA ...(((((.((..(((..(((((((..(((((((((((...((((........((((((.((....))))))))))))....)))))))))))...-)))))))...)))..)).))))) ( -32.40) >DroPer_CAF1 2 108 + 1 -----------AAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU-UUUGCGUUGACCUCGUUUACGGGUCAGAGCGGGUAUGAAAAGUUUGCCCGUACAAACUAAAUUGCCGCCUGCA -----------.....((((..((((.(((.....)))..))))..-...))))(((((.((....)))))))..((((((......((((((......))))))........)))))). ( -26.14) >consensus CGGUGCAACUGAAAAUACGUUUGCAUAUGAAUUUUUCAAAAUGUUU_UUUCCGCUGACCUCGUUUACGGGUCAGAGCAGCUAUGAAAAGUUUGCCC_UGCAAACUAAAUUGCCGUUUGCA ..................(((((((..(((((((((((...((((........((((((.((....))))))))))))....)))))))))))....))))))).....(((.....))) (-24.72 = -24.64 + -0.08)

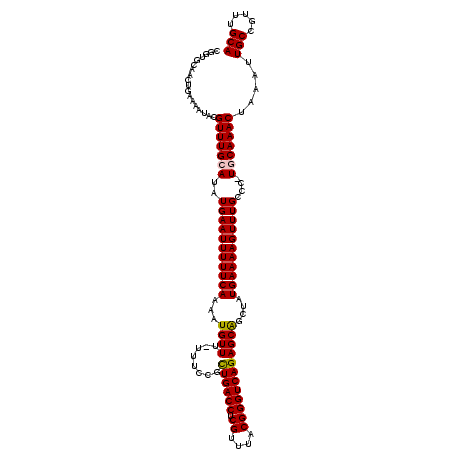

| Location | 20,697,246 – 20,697,364 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20697246 118 - 23771897 UGCAAACGGCAAUUUAGUUUGCA-GGGCAAACUUUUCAUAGCUGCUCUGACCCGUAAACGAGGUCAGCGGAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCG (((((((.........)))))))-((((((...........(((((..((((((....)).)))))))))...-.......((((((.....(((((....))))))))))))))).)). ( -30.70) >DroPse_CAF1 2 108 - 1 UGCAGGCGGCAAUUUAGUUUGUACGGGCAAACUUUUCAUACCCGCUCUGACCCGUAAACGAGGUCAACGCAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUU----------- (((((((((......(((((((....)))))))........))))).(((((((....)).)))))...((((-(....)))))...........))))..........----------- ( -25.84) >DroEre_CAF1 19263 118 - 1 UGCAAACGGCAAUUUAGUUUGCA-GGGCAAACUUUUCAUAGCUGCUCUGACCCGUAAACGAGGUCAGUGGAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCG (((((((.........)))))))-((((((..((((((........((((((((....)).))))))))))))-.......((((((.....(((((....))))))))))))))).)). ( -29.80) >DroYak_CAF1 89 119 - 1 UGCAAACGGCAAUUUAGUUUGCA-GGGCAAACUUUUCAUAGCUGCUCUGACCCGUAAACGAGGUCAGCGGAAAAAAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCG (((((((.........)))))))-((((((...........(((((..((((((....)).)))))))))...........((((((.....(((((....))))))))))))))).)). ( -30.70) >DroPer_CAF1 2 108 - 1 UGCAGGCGGCAAUUUAGUUUGUACGGGCAAACUUUUCAUACCCGCUCUGACCCGUAAACGAGGUCAACGCAAA-AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUU----------- (((((((((......(((((((....)))))))........))))).(((((((....)).)))))...((((-(....)))))...........))))..........----------- ( -25.84) >consensus UGCAAACGGCAAUUUAGUUUGCA_GGGCAAACUUUUCAUAGCUGCUCUGACCCGUAAACGAGGUCAGCGGAAA_AAACAUUUUGAAAAAUUCAUAUGCAAACGUAUUUUCAGUUGCACCG ((((..((((.((..(((((((....)))))))....)).))))(.((((((((....)).)))))).)..........................))))..................... (-23.78 = -24.10 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:11 2006