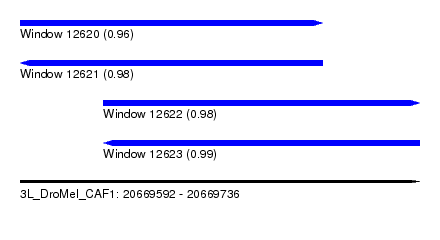
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,669,592 – 20,669,736 |
| Length | 144 |
| Max. P | 0.986378 |

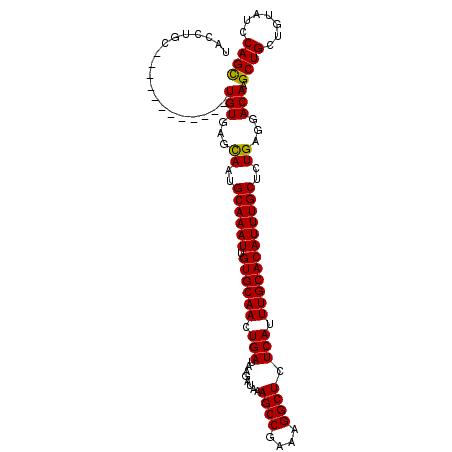
| Location | 20,669,592 – 20,669,701 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -27.97 |
| Energy contribution | -27.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20669592 109 + 23771897 UACCUGAGAGAGUAAUGUGUGUGAGUAAUGCAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUAUUUCCAGU ..(((.((((...((((((((((((...((((.....)))).............((((....)))))))))..)))))))..)))).)))....((((.......)))) ( -36.50) >DroSec_CAF1 41531 97 + 1 UACCUGC------------UGUGAGCAAUGCAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGC .....((------------(((...((..((((((.((((((.(((........((((....)))).))).))))))))))))..))...)).))).((((....)))) ( -31.50) >DroSim_CAF1 47432 97 + 1 UACCUGC------------UGUGAGCAAUGCAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGC .....((------------(((...((..((((((.((((((.(((........((((....)))).))).))))))))))))..))...)).))).((((....)))) ( -31.50) >consensus UACCUGC____________UGUGAGCAAUGCAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGC ...................(((...((..((((((.((((((.(((........((((....)))).))).))))))))))))..))...))).((((.......)))) (-27.97 = -27.53 + -0.44)

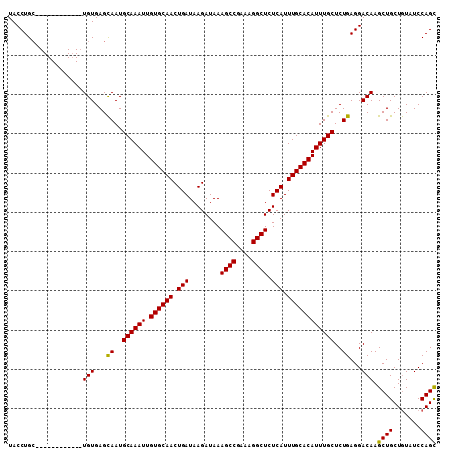
| Location | 20,669,592 – 20,669,701 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20669592 109 - 23771897 ACUGGAAAUAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUGCAUUACUCACACACAUUACUCUCUCAGGUA .((((..((((....))))..)))).((((((((((((.((((((((....))))).......))).)))))).))))))...............((((......)))) ( -25.21) >DroSec_CAF1 41531 97 - 1 GCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUGCAUUGCUCACA------------GCAGGUA ((((....)))).((((((...(((.((((((((((((.((((((((....))))).......))).)))))).)))))).)))......------------)))))). ( -33.91) >DroSim_CAF1 47432 97 - 1 GCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUGCAUUGCUCACA------------GCAGGUA ((((....)))).((((((...(((.((((((((((((.((((((((....))))).......))).)))))).)))))).)))......------------)))))). ( -33.91) >consensus GCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUGCAUUGCUCACA____________GCAGGUA ((((....)))).(((((((....))((((((((((((.((((((((....))))).......))).)))))).)))))).......................))))). (-27.93 = -28.04 + 0.11)

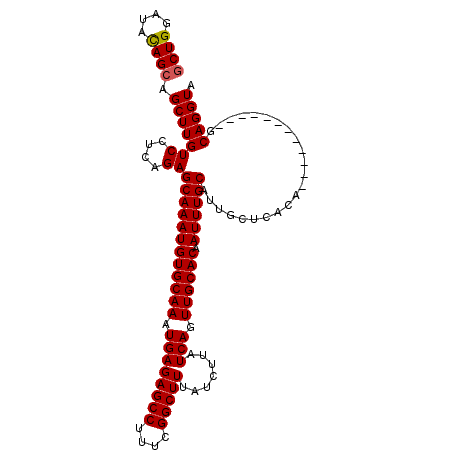

| Location | 20,669,622 – 20,669,736 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -33.79 |
| Energy contribution | -33.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20669622 114 + 23771897 CAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUAUUUCCAGUCAUCGGAAAUUCGAUGAAAGGAAGUUCAGUUUCCC (((((.((((((.(((........((((....)))).))).)))))))))))......(((..(((((..((((((..(((((((....)))))))..)))))).)))))))). ( -41.30) >DroSec_CAF1 41549 114 + 1 CAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGCCAUCGGAAAUUCGAUGAAAGGAAGUUCAGUUUCCC (((((.((((((.(((........((((....)))).))).)))))))))))......(((..(((((..(..(((...((((((....))))))...)))..).)))))))). ( -33.50) >DroSim_CAF1 47450 114 + 1 CAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGCCAUCGGAAAUUCGAUGAAAGGAAGUUCAGUUUCCA (((((.((((((.(((........((((....)))).))).)))))))))))......(((..(((((..(..(((...((((((....))))))...)))..).)))))))). ( -34.40) >consensus CAAAUUGUGCAACUGAUAAGAUAAAGCCGAAAGGCUCUCAUUUGCACAUUUGCUCUGAGGACAAGCUGCUGUAUCCAGCCAUCGGAAAUUCGAUGAAAGGAAGUUCAGUUUCCC (((((.((((((.(((........((((....)))).))).)))))))))))......(((..(((((..(..(((...((((((....))))))...)))..).)))))))). (-33.79 = -33.57 + -0.22)


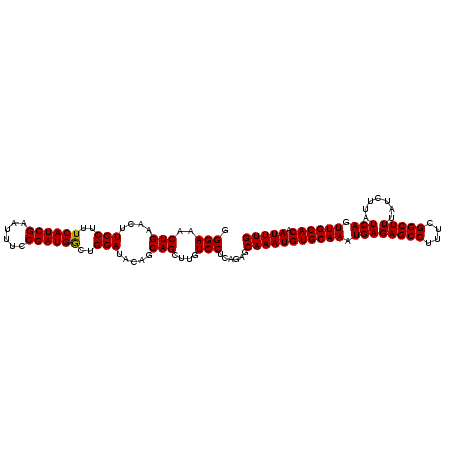
| Location | 20,669,622 – 20,669,736 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -34.70 |
| Energy contribution | -34.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20669622 114 - 23771897 GGGAAACUGAACUUCCUUUCAUCGAAUUUCCGAUGACUGGAAAUAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUG ((((..(((...((((..((((((......))))))..))))....)))....)))).....(((((((((((.((((((((....))))).......))).)))))).))))) ( -36.21) >DroSec_CAF1 41549 114 - 1 GGGAAACUGAACUUCCUUUCAUCGAAUUUCCGAUGGCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUG ((((..(((....(((..((((((......))))))..))).....)))....)))).....(((((((((((.((((((((....))))).......))).)))))).))))) ( -34.91) >DroSim_CAF1 47450 114 - 1 UGGAAACUGAACUUCCUUUCAUCGAAUUUCCGAUGGCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUG .(((..(((....(((..((((((......))))))..))).....)))....)))......(((((((((((.((((((((....))))).......))).)))))).))))) ( -34.41) >consensus GGGAAACUGAACUUCCUUUCAUCGAAUUUCCGAUGGCUGGAUACAGCAGCUUGUCCUCAGAGCAAAUGUGCAAAUGAGAGCCUUUCGGCUUUAUCUUAUCAGUUGCACAAUUUG .(((..(((....(((..((((((......))))))..))).....)))....)))......(((((((((((.((((((((....))))).......))).)))))).))))) (-34.70 = -34.48 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:03 2006