
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,648,577 – 20,648,682 |
| Length | 105 |
| Max. P | 0.706331 |

| Location | 20,648,577 – 20,648,682 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -12.23 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
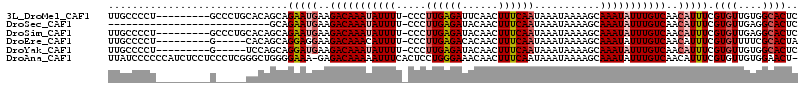
>3L_DroMel_CAF1 20648577 105 + 23771897 UUGCCCCU---------GCCCUGCACAGCAGAAUGAAGACAAAUAUUUU-CCCUUGAGAUUCAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGCACUC .((((...---------((...))(((((((((((..(((((((((((.-...((((((......))))))...........)))))))))))..)))))..))))))))))... ( -24.36) >DroSec_CAF1 21326 87 + 1 ---------------------------GCAGAAUGAAGACAAAUAUUUU-CCCUUGAGAUACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGAGGCACUC ---------------------------...(((((..(((((((((((.-...((((((......))))))...........)))))))))))..))))).((((....)))).. ( -16.76) >DroSim_CAF1 24604 105 + 1 UUGCCCCU---------GCCCUGCACAGCAGAAUGAAGACAAAUAUUUU-CCCUUGAGAUACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGAGGCACUC .((((..(---------((...)))((((((((((..(((((((((((.-...((((((......))))))...........)))))))))))..)))))..))))).))))... ( -23.96) >DroEre_CAF1 10190 100 + 1 UUGCCCCU---------G-----CACAGCAGGAGGAAGACAAACAUUUU-CCCUUGAGACACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUUUCGCACUA .(((.(((---------(-----(...))))).((((((......))))-))...(((((((........(((((((........))))))).........))))))).)))... ( -19.13) >DroYak_CAF1 201451 100 + 1 UUGCCCCU---------G-----UCCAGCAGGAUGAAGACAAAUAUUUU-CCCUUGAGAUACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGCACUC .......(---------(-----(((((((.((((..(((((((((((.-...((((((......))))))...........)))))))))))..))..)).))))).))))... ( -22.86) >DroAna_CAF1 21268 113 + 1 UUAUCCCCCCAUCUCCUCCCUCGGGCUGGGGAAA-GAGACAAAAAUUUCACUCCUGGGAAACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGAACU- ...(((.(((((((((.((....))..)))))..-((((......)))).....))))..(((((.....(((((((........)))))))..((.....))))))))))...- ( -22.40) >consensus UUGCCCCU_________G_____CACAGCAGAAUGAAGACAAAUAUUUU_CCCUUGAGAUACAACUUUCAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGCACUC ..............................(((((..(((((((((((.....((((((......))))))...........)))))))))))..))))).((((....)))).. (-12.23 = -13.04 + 0.81)
| Location | 20,648,577 – 20,648,682 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -14.65 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
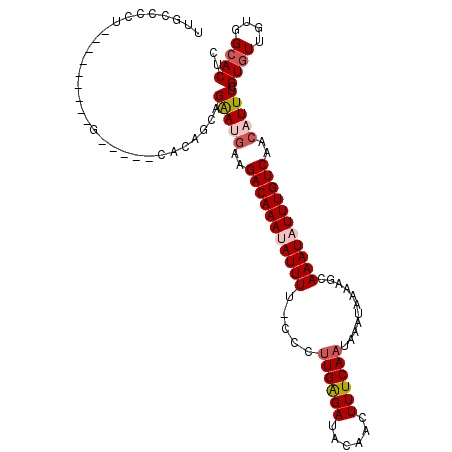
>3L_DroMel_CAF1 20648577 105 - 23771897 GAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGAAUCUCAAGGG-AAAAUAUUUGUCUUCAUUCUGCUGUGCAGGGC---------AGGGGCAA ...((((.........((((..((((((((((((((((((.....)))))))....(((....))-))))))))))))..))))(((((......)))---------)).)))). ( -30.70) >DroSec_CAF1 21326 87 - 1 GAGUGCCUCAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUAUCUCAAGGG-AAAAUAUUUGUCUUCAUUCUGC--------------------------- ..(((......)))(((.((..((((((((((((((((((.....)))))))....(((....))-))))))))))))..)))))...--------------------------- ( -20.40) >DroSim_CAF1 24604 105 - 1 GAGUGCCUCAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUAUCUCAAGGG-AAAAUAUUUGUCUUCAUUCUGCUGUGCAGGGC---------AGGGGCAA ...((((((.......((((..((((((((((((((((((.....)))))))....(((....))-))))))))))))..))))((((...))))...---------.)))))). ( -31.80) >DroEre_CAF1 10190 100 - 1 UAGUGCGAAAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUGUCUCAAGGG-AAAAUGUUUGUCUUCCUCCUGCUGUG-----C---------AGGGGCAA ..(((......)))........((((((((((((((((((.....)))))))....(((....))-))))))))))))...(((((((...)-----)---------)))))... ( -27.40) >DroYak_CAF1 201451 100 - 1 GAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUAUCUCAAGGG-AAAAUAUUUGUCUUCAUCCUGCUGGA-----C---------AGGGGCAA ...((((..........(((..((((((((((((((((((.....)))))))....(((....))-))))))))))))..)))((((.....-----)---------))))))). ( -28.20) >DroAna_CAF1 21268 113 - 1 -AGUUCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUUUCCCAGGAGUGAAAUUUUUGUCUC-UUUCCCCAGCCCGAGGGAGGAGAUGGGGGGAUAA -.(((((....(((.(((((((....)))))))(((((((.....)))))))............))).....((..((((-(((((.(.....).)))))))))..))))))).. ( -29.70) >consensus GAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUGAAAGUUGUAUCUCAAGGG_AAAAUAUUUGUCUUCAUUCUGCUGUG_____C_________AGGGGCAA ..((.((..((((.....))))((((((((((((((((((.....)))))))....((....))...)))))))))))..............................)).)).. (-14.65 = -15.18 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:41 2006