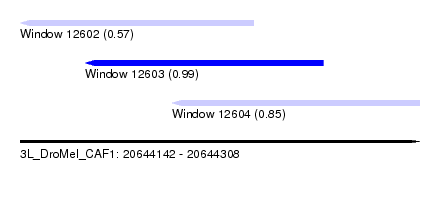
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,644,142 – 20,644,308 |
| Length | 166 |
| Max. P | 0.988358 |

| Location | 20,644,142 – 20,644,239 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -19.16 |
| Consensus MFE | -17.70 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20644142 97 - 23771897 UUAUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAAAAAAACGAGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUAAAAAUCUCGG ....((((((((((.................))))))..((((((((((....))))))))))...))))...........(((((....))))).. ( -21.43) >DroSec_CAF1 17261 95 - 1 ACAUGCGUUUGUGUGUAGACAAAAUAAAAAAACACAA--AAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUAAAAAUCUCGG .(((.(((((.(((((...............))))).--))))).((((((....))))))(((((((...))))))))))(((((....))))).. ( -20.56) >DroSim_CAF1 20564 95 - 1 ACAUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAA--AAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUAAAAAUCUCGG .(((.(((((.(((((...............))))).--))))).((((((....))))))(((((((...))))))))))(((((....))))).. ( -20.56) >DroEre_CAF1 6112 93 - 1 --AUGCGUUUGUGUGUAGACAAAAUAGAAAAAUACAA-AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUA-AAAUCUCGG --..((((((((((.................))))))-.((((((((((....))))))))))...))))...........(((((.-..))))).. ( -17.53) >DroYak_CAF1 197256 94 - 1 --AUGCGUUUGAAUGUAGACAAAAUAGAAAAAAUCAA-AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUACAAAUCUCGG --...((((..((.(..(.(......((.....))..-.((((((((((....)))))))))).....))..)))..))))(((((....))))).. ( -16.90) >DroAna_CAF1 16709 92 - 1 --AUGCGUUAGUGUGUGGACAAAAUAAAA-UAUAAA--AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUAUGAAUCGAGAUAAAUAUCUUGA --..(((((..(((((............)-))))..--.((((((((((....))))))))))..))))).........(((((((....))))))) ( -18.00) >consensus __AUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAA_AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGCCGCUUUGAAUGGAGAUAAAAAUCUCGG ....((((((((((.................))))....((((((((((....)))))))))).))))))...........(((((....))))).. (-17.70 = -17.48 + -0.22)

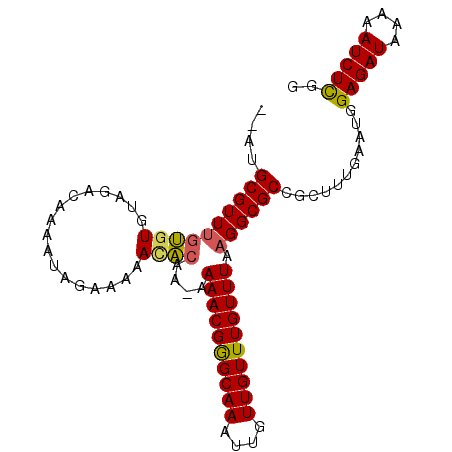
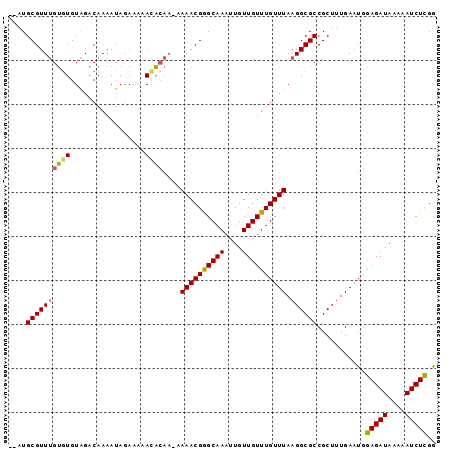
| Location | 20,644,169 – 20,644,268 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20644169 99 - 23771897 UGGCACGCAAAUGUAUCUUCAAGAUACAUUUAUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAAAAAAACGAGCAAAUUGUUGUUUGUUUAAGGCGC ..(((((((((((((((.....))))))))..)))))((((((.................))))))..((((((((((....))))))))))...)).. ( -27.53) >DroSec_CAF1 17288 97 - 1 UGGCACGCAAAUGUAUCUUUACGAUACAUACAUGCGUUUGUGUGUAGACAAAAUAAAAAAACACAA--AAACGGGCAAAUUGUUGUUUGUUUAAGGCGC ..(((((((.(((((((.....)))))))...)))))((((((.................))))))--((((((((((....))))))))))...)).. ( -25.13) >DroSim_CAF1 20591 97 - 1 UGGCACGCAAAUGUAUCUUCAAGAUACAUACAUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAA--AAACGGGCAAAUUGUUGUUUGUUUAAGGCGC ..(((((((.(((((((.....)))))))...)))))((((((.................))))))--((((((((((....))))))))))...)).. ( -25.43) >DroEre_CAF1 6138 94 - 1 UGGCACGCAAAUGUAUCUUCGAGAUAC----AUGCGUUUGUGUGUAGACAAAAUAGAAAAAUACAA-AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGC ..((((((((((((((...........----)))))))))))))).....................-.((((((((((....))))))))))....... ( -23.20) >DroYak_CAF1 197283 94 - 1 UGGCACGCAAAUGUAUCUUCAAGAUAC----AUGCGUUUGAAUGUAGACAAAAUAGAAAAAAUCAA-AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGC ..(((((((..((((((.....)))))----)))))).............................-.((((((((((....))))))))))...)).. ( -20.70) >DroAna_CAF1 16736 92 - 1 UGGCACGCAAAUGUAUCUUCAAGAUAC----AUGCGUUAGUGUGUGGACAAAAUAAAA-UAUAAA--AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGC (.((((((..(((((((.....)))))----))......)))))).)...........-......--.((((((((((....))))))))))....... ( -20.90) >consensus UGGCACGCAAAUGUAUCUUCAAGAUAC____AUGCGUUUGUGUGUAGACAAAAUAGAAAAACACAA_AAAACGGGCAAAUUGUUGUUUGUUUAAGGCGC ..((((((((((((((...............)))))))))))))).......................((((((((((....))))))))))....... (-20.97 = -21.33 + 0.36)

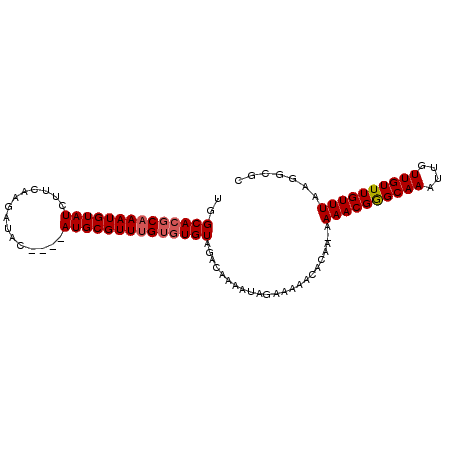
| Location | 20,644,205 – 20,644,308 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -10.17 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20644205 103 - 23771897 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUAUCUUCAA-----------GAUACAUUUAUGCGUUUGUGUGUAGACAAAAUAGAAAAACA- ....(((((((.......(((((.((((.......)))))))))(((((((((((((.....-----------))))))))..))))))))))))...................- ( -27.40) >DroPse_CAF1 23451 88 - 1 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACUCCAUUGGCACGCAAAUGUAUCUUGAA----------CAAUACAAAUA----------------C-AAAUACAAAUACAA ..(((..(((.......)))..(((((.........)))))))).......(((((.(((..----------.....))))))----------------)-)............. ( -9.90) >DroSec_CAF1 17322 103 - 1 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUAUCUUUAC-----------GAUACAUACAUGCGUUUGUGUGUAGACAAAAUAAAAAAACA- ....(((((((.......(((((.((((.......)))))))))(((((.(((((((.....-----------)))))))...))))))))))))...................- ( -25.90) >DroWil_CAF1 11539 109 - 1 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUGUCUUGAUAUAAAUUGAAAAAAAUAUAUAUACAUAGGUAUAUAAA--AAUGAGGAAA---- ...((.(((((((..(((((((((((((.......))))))((((((....)))))).)))))))))))).........(((((((....)))))))..--..)).))...---- ( -23.20) >DroYak_CAF1 197318 99 - 1 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUAUCUUCAA-----------GAUAC----AUGCGUUUGAAUGUAGACAAAAUAGAAAAAAU- ..(((((..(((......)))..)))))...(((((((...)))(((((..((((((.....-----------)))))----))))))......))))................- ( -22.10) >DroAna_CAF1 16770 98 - 1 UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUAUCUUCAA-----------GAUAC----AUGCGUUAGUGUGUGGACAAAAUAAAA-UAUA- ..((((((((.(((((.((((((.((((.......)))))))))).(((..((((((.....-----------)))))----)))))))))))))))).).........-....- ( -28.70) >consensus UAGCCGCACAAUUGAUAUGUUAAGUGGUAAAUUACGCCAUUGGCACGCAAAUGUAUCUUCAA___________GAUACAUAUAUGCGUUUGUGUGUAGACAAAAUAGAAAAACA_ .......(((.(((...((((((.((((.......))))))))))..))).)))............................................................. (-10.17 = -10.33 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:38 2006