
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,636,642 – 20,636,743 |
| Length | 101 |
| Max. P | 0.929748 |

| Location | 20,636,642 – 20,636,743 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
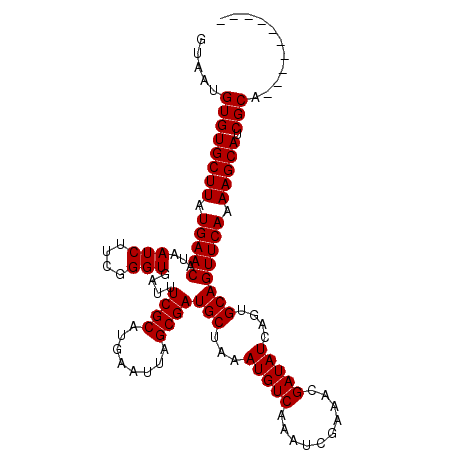
>3L_DroMel_CAF1 20636642 101 + 23771897 GUAAUGUGUGCUUAUGAACAUAAUCUUCGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAACGAUAUCAGUGCAGUUCAAAAGCAUCGCA---------- ....(((((((((.(((((...(((....)))(((((.((((...(((((...))))))))).))))).................))))).))))).))))---------- ( -27.30) >DroPse_CAF1 16314 111 + 1 GUAAUGUGUGCUUAUGAACAUAAUCCGCGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAAUGAUAUCAGUGCAGUUCAAAAGCAUCGCCUCGACGUGGA ....(((((........))))).((((((.(((....)))........((((((((..((((((........))))))..(.......)...)))))))).....)))))) ( -28.10) >DroSim_CAF1 11534 101 + 1 GUAAUGUGUGCUUAUGAACAUAAUCUUCGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAACGAUAUCAGUGCAGUUCAAAAGCAUCGCA---------- ....(((((((((.(((((...(((....)))(((((.((((...(((((...))))))))).))))).................))))).))))).))))---------- ( -27.30) >DroYak_CAF1 189803 101 + 1 GUAAUGUGUGCUUAUGAACAUAAUCUUCUGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAACGAAACGAUAUCAGUGCAGUUCAAAAGCAUCGCA---------- ....(((((((((.(((((........((((((.(((((..(((.(((((...)))))...)))...)))))...))))))....))))).))))).))))---------- ( -25.70) >DroAna_CAF1 9386 101 + 1 GUAAUGUGUGCUUAUGAACAUAAUCUUUGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAACGAUAUCAGUGCAGUUCAAAAGCAUCGCG---------- .....((((((((.(((((..((((.......))))..((((((.(((((...)))))...))..((((...))))....)))).))))).))))).))).---------- ( -24.60) >DroPer_CAF1 15205 111 + 1 GUAAUGUGUGCUUAUGAACAUAAUCCGCGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAAUGAUAUCAGUGCAGUUCAAAAGCAUCGCCUCGACGUGGA ....(((((........))))).((((((.(((....)))........((((((((..((((((........))))))..(.......)...)))))))).....)))))) ( -28.10) >consensus GUAAUGUGUGCUUAUGAACAUAAUCUUCGGGUGAUUUCGCAUGAAUUAGCGAUGCUAAAUGUCAAAUCGAAACGAUAUCAGUGCAGUUCAAAAGCAUCGCA__________ .....((((((((.(((((.................((((........))))(((...(((((..........)))))....)))))))).))))).)))........... (-24.20 = -24.20 + -0.00)


| Location | 20,636,642 – 20,636,743 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.72 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20636642 101 - 23771897 ----------UGCGAUGCUUUUGAACUGCACUGAUAUCGUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCGAAGAUUAUGUUCAUAAGCACACAUUAC ----------((.(.(((((.(((((....((......((((((...(((...(((((...))))).)))..)))))).......)).....))))).)))))).)).... ( -21.82) >DroPse_CAF1 16314 111 - 1 UCCACGUCGAGGCGAUGCUUUUGAACUGCACUGAUAUCAUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCGCGGAUUAUGUUCAUAAGCACACAUUAC ....(((....))).(((((.(((((....(((.....((((((...(((...(((((...))))).)))..))))))......))).....))))).)))))........ ( -25.70) >DroSim_CAF1 11534 101 - 1 ----------UGCGAUGCUUUUGAACUGCACUGAUAUCGUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCGAAGAUUAUGUUCAUAAGCACACAUUAC ----------((.(.(((((.(((((....((......((((((...(((...(((((...))))).)))..)))))).......)).....))))).)))))).)).... ( -21.82) >DroYak_CAF1 189803 101 - 1 ----------UGCGAUGCUUUUGAACUGCACUGAUAUCGUUUCGUUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCAGAAGAUUAUGUUCAUAAGCACACAUUAC ----------((.(.(((((.(((((....((......(((((((..(((...(((((...))))).))).))))))).......)).....))))).)))))).)).... ( -22.82) >DroAna_CAF1 9386 101 - 1 ----------CGCGAUGCUUUUGAACUGCACUGAUAUCGUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCAAAGAUUAUGUUCAUAAGCACACAUUAC ----------.(.(.(((((.(((((....((......((((((...(((...(((((...))))).)))..)))))).......)).....))))).)))))).)..... ( -21.62) >DroPer_CAF1 15205 111 - 1 UCCACGUCGAGGCGAUGCUUUUGAACUGCACUGAUAUCAUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCGCGGAUUAUGUUCAUAAGCACACAUUAC ....(((....))).(((((.(((((....(((.....((((((...(((...(((((...))))).)))..))))))......))).....))))).)))))........ ( -25.70) >consensus __________UGCGAUGCUUUUGAACUGCACUGAUAUCGUUUCGAUUUGACAUUUAGCAUCGCUAAUUCAUGCGAAAUCACCCGAAGAUUAUGUUCAUAAGCACACAUUAC ...........(.(.(((((.(((((....((......((((((...(((...(((((...))))).)))..)))))).......)).....))))).)))))).)..... (-21.16 = -20.72 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:31 2006