
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,634,777 – 20,634,879 |
| Length | 102 |
| Max. P | 0.893878 |

| Location | 20,634,777 – 20,634,879 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -20.24 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
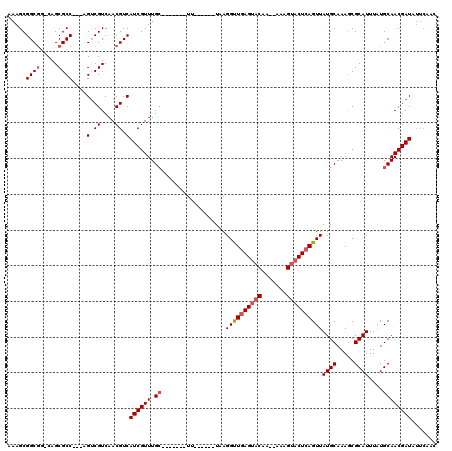
>3L_DroMel_CAF1 20634777 102 + 23771897 AAAGCGGCGGACAGCGCC---AGUCGUCAACGUCAUCGUUUGC-------UU------UAAGGCUGAGUACAA--AAAGUACUCAGUUAUGCAAAGCGCAUUUAUGCAACGAUAUUCAAC .....((((.....))))---.(.((....)).)((((((.((-------((------(..((((((((((..--...))))))))))....)))))(((....)))))))))....... ( -33.10) >DroSec_CAF1 7953 100 + 1 AAAGCGGCGG-CAGCGCC---AGUCGUCAACGUCAUCGUUUGC-------UU------UGAGGUUGAGUACAA---AAGUACUCAGUUAUGCAAAGCGCAUUUAUGCAACGAUAUUCAAC ...(((((((-(...)))---.))))).......((((((.((-------((------((.(..(((((((..---..)))))))..)...))))))(((....)))))))))....... ( -32.50) >DroSim_CAF1 9634 100 + 1 AAAGCGGCGG-CAGCGCC---AGUCGUCAACGUCAUCGUUUGC-------UU------UGAGGUUGAGUACAA---AAGUACUCAGUUAUGCAAAGCGCAUUUAUGCAACGAUAUUCAAC ...(((((((-(...)))---.))))).......((((((.((-------((------((.(..(((((((..---..)))))))..)...))))))(((....)))))))))....... ( -32.50) >DroYak_CAF1 187933 103 + 1 AAAGCGGCGG-CAGCGCC---AGUCGUCAACGUCAUCGUUUGC-------UU------UAAGGUUGAGUACAAAAAAAGUACUCAGUUAUGCAAAGCGCAUUUAUGCAACGAUAUUCACC ...(((((((-(...)))---.))))).......((((((.((-------((------(.(..((((((((.......))))))))...)..)))))(((....)))))))))....... ( -28.20) >DroAna_CAF1 7779 110 + 1 AAAGCGGCAC-CAGCGCCGUCCGCCGUCAACGUCAUCGUUUGACUUUAGCUUUGGCCAUAAGGUUCAGGCCAG--AAAGUACUCAGUUAUGCAAAACGCAUUUGUGCAACGAU------- ...(((((..-....)))))..((.(((((((....)).)))))....))(((((((..........))))))--)..((((......((((.....))))..))))......------- ( -29.40) >consensus AAAGCGGCGG_CAGCGCC___AGUCGUCAACGUCAUCGUUUGC_______UU______UAAGGUUGAGUACAA__AAAGUACUCAGUUAUGCAAAGCGCAUUUAUGCAACGAUAUUCAAC .....((((.....))))....(.((....)).)((((((.((..................((((((((((.......))))))))))((((.....))))....))))))))....... (-20.24 = -21.08 + 0.84)


| Location | 20,634,777 – 20,634,879 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
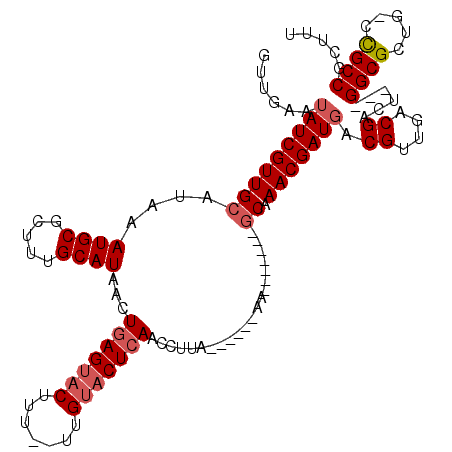
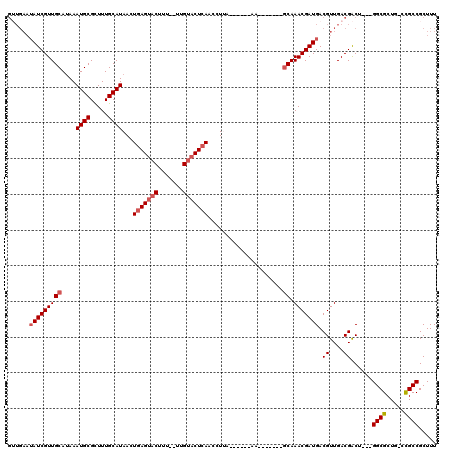
>3L_DroMel_CAF1 20634777 102 - 23771897 GUUGAAUAUCGUUGCAUAAAUGCGCUUUGCAUAACUGAGUACUUU--UUGUACUCAGCCUUA------AA-------GCAAACGAUGACGUUGACGACU---GGCGCUGUCCGCCGCUUU ((..(.((((((((((....)))((((((.....((((((((...--..))))))))...))------))-------)).)))))))...)..))....---((((.....))))..... ( -34.60) >DroSec_CAF1 7953 100 - 1 GUUGAAUAUCGUUGCAUAAAUGCGCUUUGCAUAACUGAGUACUU---UUGUACUCAACCUCA------AA-------GCAAACGAUGACGUUGACGACU---GGCGCUG-CCGCCGCUUU ((..(.((((((((((....)))((((((......(((((((..---..)))))))....))------))-------)).)))))))...)..))....---((((...-.))))..... ( -33.40) >DroSim_CAF1 9634 100 - 1 GUUGAAUAUCGUUGCAUAAAUGCGCUUUGCAUAACUGAGUACUU---UUGUACUCAACCUCA------AA-------GCAAACGAUGACGUUGACGACU---GGCGCUG-CCGCCGCUUU ((..(.((((((((((....)))((((((......(((((((..---..)))))))....))------))-------)).)))))))...)..))....---((((...-.))))..... ( -33.40) >DroYak_CAF1 187933 103 - 1 GGUGAAUAUCGUUGCAUAAAUGCGCUUUGCAUAACUGAGUACUUUUUUUGUACUCAACCUUA------AA-------GCAAACGAUGACGUUGACGACU---GGCGCUG-CCGCCGCUUU .((.((..((((((((....)))((((((......(((((((.......)))))))....))------))-------))....)))))..)).))....---((((...-.))))..... ( -29.10) >DroAna_CAF1 7779 110 - 1 -------AUCGUUGCACAAAUGCGUUUUGCAUAACUGAGUACUUU--CUGGCCUGAACCUUAUGGCCAAAGCUAAAGUCAAACGAUGACGUUGACGGCGGACGGCGCUG-GUGCCGCUUU -------...((.((((....(((((((((...............--.(((((..........)))))........(((((.((....))))))).)))))..))))..-)))).))... ( -31.40) >consensus GUUGAAUAUCGUUGCAUAAAUGCGCUUUGCAUAACUGAGUACUUU__UUGUACUCAACCUUA______AA_______GCAAACGAUGACGUUGACGACU___GGCGCUG_CCGCCGCUUU ......(((((((((....((((.....))))...(((((((.......))))))).....................)).))))))).((....))......((((.....))))..... (-22.64 = -23.48 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:29 2006