| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,621,593 – 20,621,700 |
| Length | 107 |
| Max. P | 0.677222 |

| Location | 20,621,593 – 20,621,700 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -21.82 |
| Energy contribution | -22.96 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
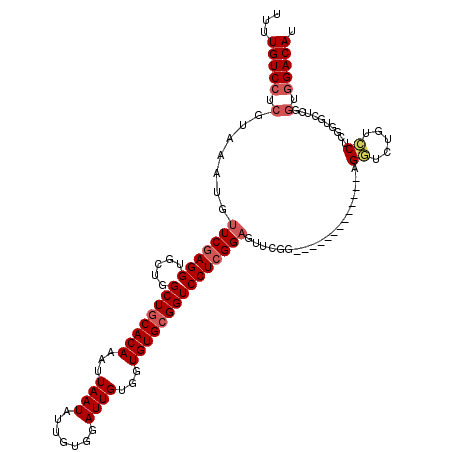
>3L_DroMel_CAF1 20621593 107 + 23771897 UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGUGGUGUGCGGUCCUCGGAGUUCGG-------------AGUUCUAUCCUCGGUGCUCGGUGGACAU ...(((((.(.........(((((.....((((((((...((((.......))))...)))))))))))))((((.(.(-------------((.......))).).)))).).))))). ( -35.30) >DroSec_CAF1 66345 107 + 1 UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGUGGUGUGCGGUCCUCGGAGUUCGG-------------AGGUCUGUCCUCCGUGCUCGGUGGACAU ...(((((.(.........(((((.....((((((((...((((.......))))...)))))))))))))((((.(((-------------(((.....)))))).)))).).))))). ( -43.70) >DroAna_CAF1 88086 83 + 1 UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGCGGUGUGAGGUCCUGGGUCCUCG-------------------------------------GACAU ...((((((((........))).((((.....))))......(((((((.....)))))))((((.(....).)))))-------------------------------------)))). ( -24.60) >DroSim_CAF1 61245 107 + 1 UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGUGGUGUGCGGUCCUCGGAGUUCGG-------------AGGUCUGUCCUCGGUGCUCGGUGGACAU ...(((((.(.........(((((.....((((((((...((((.......))))...)))))))))))))((((.(.(-------------(((.....)))).).)))).).))))). ( -39.40) >DroEre_CAF1 58799 120 + 1 UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGUGGUGUGCGGUCCUCGGAGAUCGGUCCUCGUUCCCGGUGCUCAGUGCUCAGUGCUCGGUGGACAU ...(((((..........((((.(..((((..((((..........((.(((((((.....))))))).))((((((((.........))))).))).))))))))..))))).))))). ( -39.10) >consensus UUUUGUCCUCGUAAAUGUUCGAGGUGCUGGCUGCACAAAUUAAUAUUGUGGAUUGUGGUGUGCGGUCCUCGGAGUUCGG_____________AGGUCUGUCCUCGGUGCUCGGUGGACAU ...(((((.(.......(((((((.....((((((((...((((.......))))...)))))))))))))))....................((.....))..........).))))). (-21.82 = -22.96 + 1.14)


| Location | 20,621,593 – 20,621,700 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
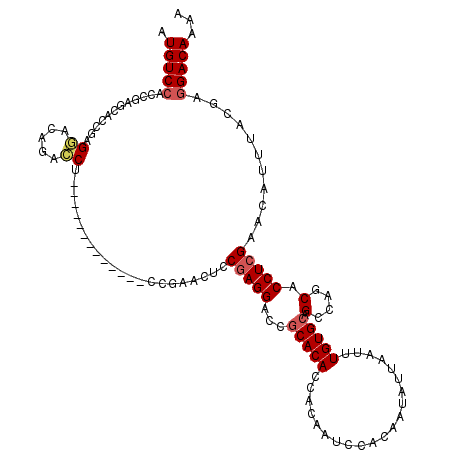
>3L_DroMel_CAF1 20621593 107 - 23771897 AUGUCCACCGAGCACCGAGGAUAGAACU-------------CCGAACUCCGAGGACCGCACACCACAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .(((((...(((..(.(((.......))-------------).)..)))(((((...(((((.....................))))).(....).)))))...........)))))... ( -24.90) >DroSec_CAF1 66345 107 - 1 AUGUCCACCGAGCACGGAGGACAGACCU-------------CCGAACUCCGAGGACCGCACACCACAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .(((((...(((..((((((.....)))-------------)))..)))(((((...(((((.....................))))).(....).)))))...........)))))... ( -33.30) >DroAna_CAF1 88086 83 - 1 AUGUC-------------------------------------CGAGGACCCAGGACCUCACACCGCAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .((((-------------------------------------(((((.(....).)))).........................((((.....)))).(((........))))))))... ( -18.90) >DroSim_CAF1 61245 107 - 1 AUGUCCACCGAGCACCGAGGACAGACCU-------------CCGAACUCCGAGGACCGCACACCACAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .(((((...(((..(.((((.....)))-------------).)..)))(((((...(((((.....................))))).(....).)))))...........)))))... ( -27.20) >DroEre_CAF1 58799 120 - 1 AUGUCCACCGAGCACUGAGCACUGAGCACCGGGAACGAGGACCGAUCUCCGAGGACCGCACACCACAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .(((((..((((..(((.((...(..(..(((((.((.....)).)).)))..)..)(((((.....................))))).)))))...))))..(......).)))))... ( -28.40) >consensus AUGUCCACCGAGCACCGAGGACAGACCU_____________CCGAACUCCGAGGACCGCACACCACAAUCCACAAUAUUAAUUUGUGCAGCCAGCACCUCGAACAUUUACGAGGACAAAA .(((((............((.....))......................(((((...(((((.....................))))).(....).)))))...........)))))... (-14.14 = -14.88 + 0.74)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:23 2006