
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,620,282 – 20,620,590 |
| Length | 308 |
| Max. P | 0.983815 |

| Location | 20,620,282 – 20,620,401 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20620282 119 + 23771897 CGAUAAUCGACCUGCCUCAUUU-CGCAAGAGUAUGUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUU ......(((....(((.(((((-(....))).))).)))....)))..................(((((((....((((........))))..)))))))...(((((....)))))... ( -30.10) >DroSec_CAF1 65031 119 + 1 CGAUAAUCGACCUGCCUCAUUU-CGCAAGAGUAUGUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUU ......(((....(((.(((((-(....))).))).)))....)))..................(((((((....((((........))))..)))))))...(((((....)))))... ( -30.10) >DroAna_CAF1 86858 119 + 1 CGAUAACCAAGCAACCUUAUUUUGGCAAGAGCAUAUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCU-CU .....(((((((.......((((((...(.((.....)))..))))))...........................((((........))))..)))))))...(((((....)))))-.. ( -24.80) >DroSim_CAF1 59942 119 + 1 CGAUAAUCGACCUGCCUCAUUU-CGCAAGAGUAUGUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUU ......(((....(((.(((((-(....))).))).)))....)))..................(((((((....((((........))))..)))))))...(((((....)))))... ( -30.10) >DroEre_CAF1 57550 119 + 1 CGAUAAUCGACCUGCCUCAUUU-GGCAAGAGUAUGUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUU .((..(((((((((((......-))).)).)).((((..(((..((................))(((((((....((((........))))..)))))))..)))..))))))))..)). ( -28.99) >consensus CGAUAAUCGACCUGCCUCAUUU_CGCAAGAGUAUGUGGCCCACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUU ......(((....(((.(((...(....)...))).)))....)))..................(((((((....((((........))))..)))))))...(((((....)))))... (-21.88 = -22.56 + 0.68)



| Location | 20,620,321 – 20,620,439 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20620321 118 + 23771897 CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGC ...................................((((........))))..(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..))))).... ( -31.10) >DroSec_CAF1 65070 118 + 1 CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGC ...................................((((........))))..(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..))))).... ( -31.10) >DroAna_CAF1 86898 112 + 1 CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCU-CUAGCAGGCAAAAGAGACUUGAGUCUUAGCCCG-------GC ..(((...................(((((((....((((........))))..)))))))...(((((....)))))-......(((....(((((....))))).)))))-------). ( -27.50) >DroSim_CAF1 59981 118 + 1 CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGC ...................................((((........))))..(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..))))).... ( -31.10) >DroEre_CAF1 57589 118 + 1 CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGC ...................................((((........))))..(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..))))).... ( -31.10) >consensus CACCGAAAAAAAUAAAUAAAUAUCACUAAACACAGGCGCAAACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG__ACUUGAGCUUGAGCUUGAGUUUGAGC ...................................((((........))))..(((((.(((.(((((....))))).))).))))).......((((((((....))))))))...... (-24.88 = -25.56 + 0.68)


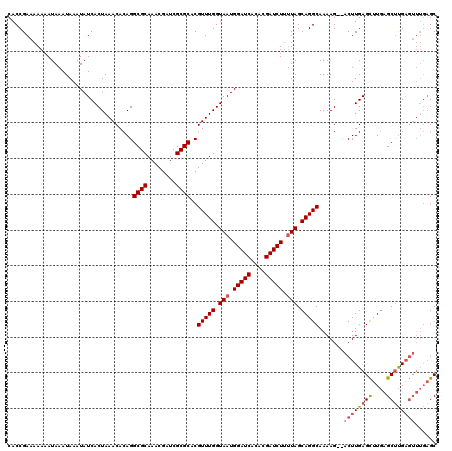
| Location | 20,620,361 – 20,620,477 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -33.08 |
| Energy contribution | -34.16 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20620361 116 + 23771897 AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACU ..(((........(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..)))))((((((..(.(((((...--.)).))).)..)))))).)))... ( -42.20) >DroSec_CAF1 65110 116 + 1 AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUAUGAGUCUGGCUCUUCGACU ..(((.((.((((...((.(((.(((((....))))).))).))(((...((--(((..(((....)))..)))))..)))..)))).))(((.--((......))..)))...)))... ( -40.60) >DroAna_CAF1 86938 112 + 1 AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCU-CUAGCAGGCAAAAGAGACUUGAGUCUUAGCCCG-------GCCAAGUGCACGGCCUGUGCCUGCAAGUCUGGGUCUUGGUCU ...((((......((.((((.....)))).))(((((-((.(((((((...(((((....))))).....(-------(((........))))..))))))).))...)))))..)))). ( -36.90) >DroSim_CAF1 60021 116 + 1 AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACU ..(((........(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..)))))((((((..(.(((((...--.)).))).)..)))))).)))... ( -42.20) >DroEre_CAF1 57629 116 + 1 AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACU ..(((........(((((.(((.(((((....))))).))).)))))...((--(((..(((....)))..)))))((((((..(.(((((...--.)).))).)..)))))).)))... ( -42.20) >consensus AACGAUCGCGCACGUUUGGUAAUGGAUCACACGAUCUUUUAGCAGGCAAAAG__ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU__GCCUGUGAGUCUGGCUCUUCGACU ...(((((((((((((((.(((.(((((....))))).))).))))).........(((.(((..(((....)))..))))))))))..(((....))).)))).))............. (-33.08 = -34.16 + 1.08)


| Location | 20,620,401 – 20,620,515 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -32.98 |
| Energy contribution | -35.26 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20620401 114 + 23771897 AGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACUGGGAUCUGGGACCCAUGCG--CAGGUCUCCACGAGGGUAU .((((((.....--(((((.(((..(((....)))..)))))))).....))))--))...........((.(((((..(((((((((.(.......).--)))))).)))))))))).. ( -45.90) >DroSec_CAF1 65150 113 + 1 AGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUAUGAGUCUGGCUCUUCGACUGGGACCUGGGAC---UGCGACUGGGUCUCCACGAGGGUAU .((((((.....--(((((.(((..(((....)))..)))))))).....))))--))...........((.(((((..(((((((..(...---.....)..)))).)))))))))).. ( -48.20) >DroAna_CAF1 86977 103 + 1 AGCAGGCAAAAGAGACUUGAGUCUUAGCCCG-------GCCAAGUGCACGGCCUGUGCCUGCAAGUCUGGGUCUUGGUCUUGC-UCUUGGUC---UUGGCCUGGGUCU------GGGUCU ..(((((....(((((....))))).(((.(-------((((((.(((.((((...((((........))))...)))).)))-.)))))))---..)))....))))------)..... ( -42.30) >DroSim_CAF1 60061 113 + 1 AGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACUGGGAUCUGGGAC---UGCGACUGGGUCUCCACGAGGGUAU .((((((.....--(((((.(((..(((....)))..)))))))).....))))--))...........((.(((((..(((((((..(...---.....)..)))).)))))))))).. ( -45.60) >DroEre_CAF1 57669 113 + 1 AGCAGGCAAAAG--ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU--GCCUGUGAGUCUGGCUCUUCGACUUGGAUCUCGGAC---UGCGACUGGGUCUCCACGAGGGUAU .((((((.....--(((((.(((..(((....)))..)))))))).....))))--))...........((.(((((...((((.(((((..---.....)))))..))))))))))).. ( -41.60) >consensus AGCAGGCAAAAG__ACUUGAGCUUGAGCUUGAGUUUGAGCCAAGUGCACGGCCU__GCCUGUGAGUCUGGCUCUUCGACUGGGAUCUGGGAC___UGCGACUGGGUCUCCACGAGGGUAU ..(((((.......(((((.(((..(((....)))..))))))))(((.(((....))))))..)))))..((((((...(((((((((...........)))))))))..))))))... (-32.98 = -35.26 + 2.28)


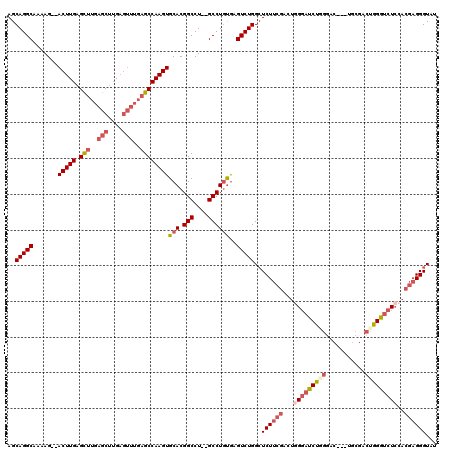
| Location | 20,620,477 – 20,620,590 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -44.06 |
| Consensus MFE | -21.18 |
| Energy contribution | -25.26 |
| Covariance contribution | 4.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20620477 113 + 23771897 GGGAUCUGGGACCCAUGCG--CAGGUCUCCACGAGGGUAUGGCCAUAGCCAGGCUAAGGCUAUGGCUA-----UGGGUCUGGCUUUGGGUAGAGCUCUGCCGGGGUAAAAUGACAUUGAC ...((((.(((((((.((.--(((..((((....)))(((((((((((((.......)))))))))))-----)))..)))))..)))))((....)).)).)))).............. ( -45.10) >DroSec_CAF1 65226 117 + 1 GGGACCUGGGAC---UGCGACUGGGUCUCCACGAGGGUAUGGCAAUAGCCAGGCUAUGGCCAUGGCUAUGGUAUGGGUCUGGCUAUGGGUAGAGCUCUGCCGGGGUAAAAUGACAUUGAC ((((((..(...---.....)..)))).)).(((...(((.((.(((((((((((...(((((....)))))...))))))))))).(((((....)))))...))...)))...))).. ( -47.80) >DroAna_CAF1 87050 98 + 1 UGC-UCUUGGUC---UUGGCCUGGGUCU------GGGUCUGGGC----UCUGGCUCAGACUCUGGCU-----CUGGCUCUGGCUAUGGGUAUGGGU---UCGGGGUAAAAUGACAUUGAC (((-((.(((((---..((((.(((((.------((((((((((----....)))))))))).))))-----).))))..))))).))))).....---((((.((......)).)))). ( -44.30) >DroSim_CAF1 60137 117 + 1 GGGAUCUGGGAC---UGCGACUGGGUCUCCACGAGGGUAUGGCCAUAGCCAGGCUAUGACCAUGGCUAUGGUAUGGGUCUGGCUAUGGGUAGAGCUCUGCCGGGGUAAAAUGACAUUGAC ((((((..(...---.....)..)))).)).(((((((.((.(((((((((((((...(((((....)))))...))))))))))))).))..)))))..)).................. ( -48.40) >DroEre_CAF1 57745 107 + 1 UGGAUCUCGGAC---UGCGACUGGGUCUCCACGAGGGUAUGGCCAUAGC-----UAUGGCUCUGG-----GUCUGGGUCUGGCCAUGGGUAAGGCUCUGCCGGGUUAAAAUGACAUUGAC ((((.(((((..---.....)))))..))))(((.((((.((((.((.(-----(((((((..((-----(......))))))))))).)).)))).))))..(((.....))).))).. ( -34.70) >consensus GGGAUCUGGGAC___UGCGACUGGGUCUCCACGAGGGUAUGGCCAUAGCCAGGCUAUGGCUAUGGCUA__GUAUGGGUCUGGCUAUGGGUAGAGCUCUGCCGGGGUAAAAUGACAUUGAC (((((((((...........))))))))).....((.((((((((((((...)))))))))))).))..........((((((...(((.....))).))))))................ (-21.18 = -25.26 + 4.08)


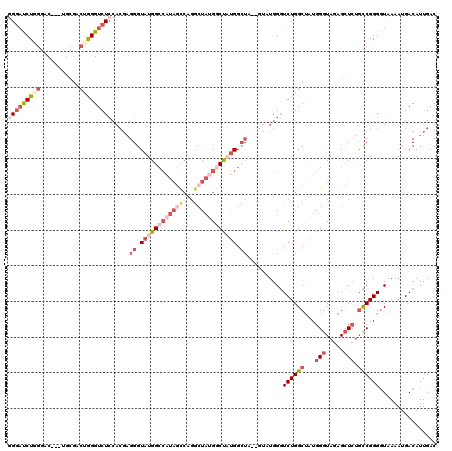
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:19 2006