
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,619,503 – 20,619,622 |
| Length | 119 |
| Max. P | 0.962142 |

| Location | 20,619,503 – 20,619,603 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.07 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20619503 100 + 23771897 GGGCUAAAAACUUAAAAGCCAGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUG .((((...........))))...........((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).))))........... ( -35.10) >DroSec_CAF1 64277 100 + 1 GGGCUAAAAACUUAAAAGCCAGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUG .((((...........))))...........((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).))))........... ( -35.10) >DroSim_CAF1 59185 100 + 1 GGGCUAAAAACUUAAAAGCCAGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUG .((((...........))))...........((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).))))........... ( -35.10) >DroEre_CAF1 56843 100 + 1 GGGCUAAAAACUUAAAAGCCAGCAAACAAAACGCUGCUCUGCAUCUUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUG .((((...........))))...........((((.(((..(((((...(((((.(.....).))))).....)).)))..))).))))........... ( -29.40) >consensus GGGCUAAAAACUUAAAAGCCAGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUG .((((...........))))...........((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).))))........... (-33.84 = -33.90 + 0.06)

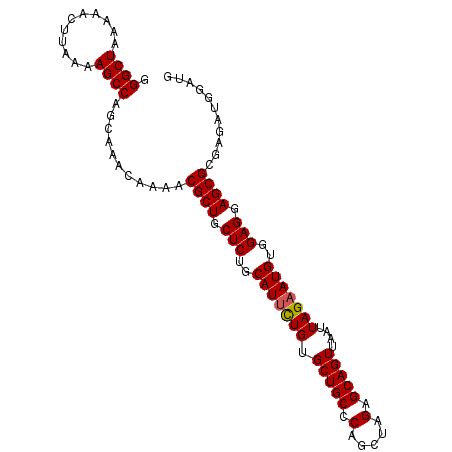
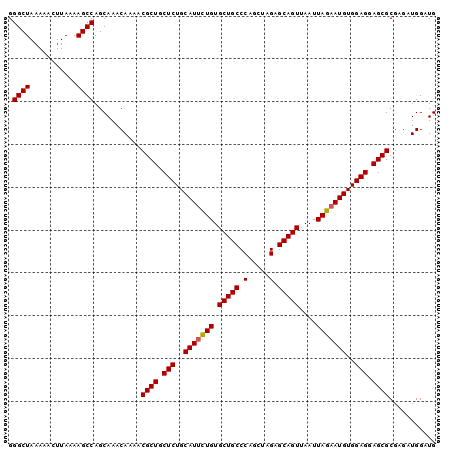
| Location | 20,619,503 – 20,619,603 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -28.21 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20619503 100 - 23771897 CAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCUGGCUUUUAAGUUUUUAGCCC ((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..))..((((...........)))). ( -29.30) >DroSec_CAF1 64277 100 - 1 CAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCUGGCUUUUAAGUUUUUAGCCC ((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..))..((((...........)))). ( -29.30) >DroSim_CAF1 59185 100 - 1 CAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCUGGCUUUUAAGUUUUUAGCCC ((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..))..((((...........)))). ( -29.30) >DroEre_CAF1 56843 100 - 1 CAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAAGAUGCAGAGCAGCGUUUUGUUUGCUGGCUUUUAAGUUUUUAGCCC ((..((....(((((.(((..(((.((......((((((.....))))))....)))))..))).)))))..))..))..((((...........)))). ( -25.00) >consensus CAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCUGGCUUUUAAGUUUUUAGCCC ((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..))..((((...........)))). (-28.21 = -28.28 + 0.06)


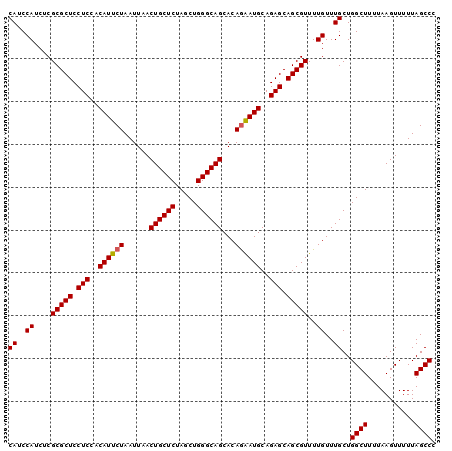
| Location | 20,619,523 – 20,619,622 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -27.40 |
| Energy contribution | -29.44 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20619523 99 + 23771897 AGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUGCUGUAUGCUGG---------------------ACCGGGAA ((((.(((....(((.(((..(((((((.(((((.(.....).)))))....)))))))..))).)))(((.......)))))).))))..---------------------........ ( -35.80) >DroSec_CAF1 64297 99 + 1 AGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUGCUGCAUGCUGG---------------------ACCGGGAA ((((..((...((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).)))).....))..)))).........---------------------........ ( -35.60) >DroAna_CAF1 86046 111 + 1 AGCAAACAAAACGCUGCUC-GCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGA--------GAGAGGGAGGCUGUGUCGCGGAUGAUGAUGGUGACGUGCCCUCCAUGCAA .(((.......(.((.(((-((((((((.(((((.(.....).)))))....))))))))))--------))).)(((((.(..(((((...........)))))..).))))).))).. ( -42.30) >DroSim_CAF1 59205 99 + 1 AGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUGCUGCAUGCUGG---------------------ACCGGGAA ((((..((...((((.(((..(((((((.(((((.(.....).)))))....)))))))..))).)))).....))..)))).........---------------------........ ( -35.60) >DroEre_CAF1 56863 94 + 1 AGCAAACAAAACGCUGCUCUGCAUCUUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUGCUG-----UGG---------------------ACCGGGCA .((........(((.((..(.(((((((((((.((((((....)).(((......))).))).).))))))))))).).)).)-----)).---------------------.....)). ( -30.04) >consensus AGCAAACAAAACGCUGCUCUGCAUUCUGUGCUGCCCAGCUAGAGCAGUUAAUUAGAAUGUGGAGGAGCGCGAGAUGGAUGCUGCAUGCUGG_____________________ACCGGGAA ((((..((...((((.((((((((((((.(((((.(.....).)))))....)))))))))))).)))).....))..))))...................................... (-27.40 = -29.44 + 2.04)

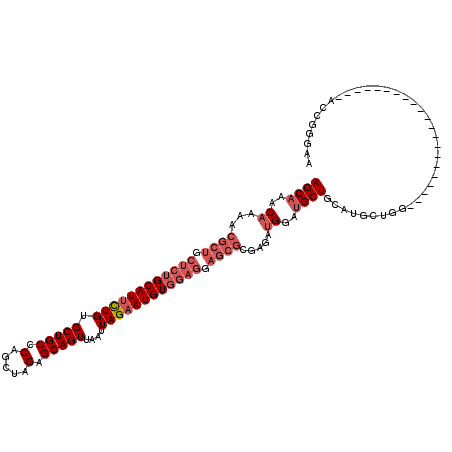
| Location | 20,619,523 – 20,619,622 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -22.68 |
| Energy contribution | -24.36 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20619523 99 - 23771897 UUCCCGGU---------------------CCAGCAUACAGCAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCU ........---------------------.........((((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..)))) ( -30.40) >DroSec_CAF1 64297 99 - 1 UUCCCGGU---------------------CCAGCAUGCAGCAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCU ........---------------------.........((((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..)))) ( -30.40) >DroAna_CAF1 86046 111 - 1 UUGCAUGGAGGGCACGUCACCAUCAUCAUCCGCGACACAGCCUCCCUCUC--------UCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGC-GAGCAGCGUUUUGUUUGCU ..(((.((..(((..(((.(...........).)))...)))..))....--------...............((((((.....)))))).(((((((((-.....))))))))).))). ( -30.10) >DroSim_CAF1 59205 99 - 1 UUCCCGGU---------------------CCAGCAUGCAGCAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCU ........---------------------.........((((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..)))) ( -30.40) >DroEre_CAF1 56863 94 - 1 UGCCCGGU---------------------CCA-----CAGCAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAAGAUGCAGAGCAGCGUUUUGUUUGCU ........---------------------...-----.((((..((....(((((.(((..(((.((......((((((.....))))))....)))))..))).)))))..))..)))) ( -26.10) >consensus UUCCCGGU_____________________CCAGCAUACAGCAUCCAUCUCGCGCUCCUCCACAUUCUAAUUAACUGCUCUAGCUGGGCAGCACAGAAUGCAGAGCAGCGUUUUGUUUGCU ......................................((((..((....(((((.(((..((((((......((((((.....))))))...))))))..))).)))))..))..)))) (-22.68 = -24.36 + 1.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:13 2006