
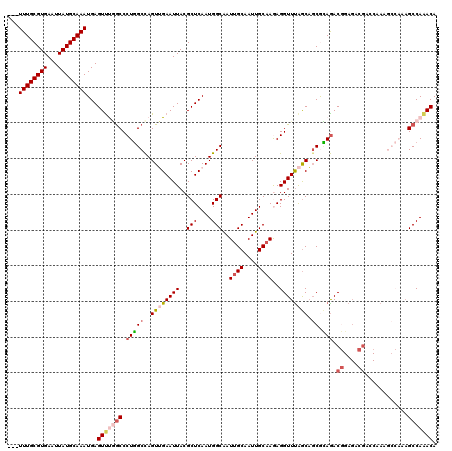
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,618,248 – 20,618,365 |
| Length | 117 |
| Max. P | 0.764597 |

| Location | 20,618,248 – 20,618,365 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -21.43 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
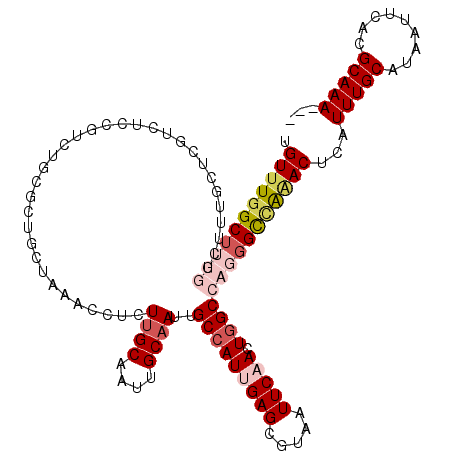
>3L_DroMel_CAF1 20618248 117 + 23771897 ---UUUGCGUGAAUUAUGCAAAUGAGUUUGGCCCUGGCUAGUUGAAUUACGCUCAAUGGCGAUUGCAAUUGCAAGAGGUUUAGCAGCGCAAACGGAGACGACCAAAGCCAAAGCCAAACA ---((((((((...))))))))...(((((((..(((((.((((((((.((((....)))).((((....))))..))))))))........((....)).....)))))..))))))). ( -41.60) >DroSec_CAF1 62987 117 + 1 ---UUUGCGUGAAUUAUGCAAAUGAGUUCGGCCCUGGCCAGUUGAAUUACGCUCAAUGGCAAUUGCAAUUGCAAGAGGUUUAGCAGCGCAGACGGAGACGACCAAAGCCAAAGCCAAACA ---((((((((...))))))))...(((.(((..((((..((((....((.(((....(((((....)))))..)))))....)))).....((....))......))))..))).))). ( -35.20) >DroAna_CAF1 84677 102 + 1 GACUUUGCGUGAAUUAUGCAAAUGAGUCUUACCCUCGCCAGUGGAAUUACGCUCAAUGGCAAUUGCAAUUGCAAAAGGUUCAACCGCAGAGAC------------------AGCCAAACA ..(((((((((((((.(((((..(((.......)))(((((((......)))....))))........)))))...))))))..)))))))..------------------......... ( -26.80) >DroSim_CAF1 57892 117 + 1 ---UUUGCGUGAAUUAUGCAAAUGAGUUCGGCCCUGGCCAGUUGAAUUACGCUCAAUGGCAAUUGCAAUUGCAAGAGGUUUAGCAGCGCAGACGGAGACGAGCAAAGCCAAAGCCAAACA ---((((((((...))))))))...(((.(((..((((..((((....((.(((....(((((....)))))..)))))....))))((...((....)).))...))))..))).))). ( -36.30) >DroEre_CAF1 55572 111 + 1 ---UUUGCGUGAAUUAUGCAAAUGAGUUUAGCCCUGGCCAGUAGAAUUACGCUCAAUGGCAAUCGCAAUUGCCAAAGGUUUGGCAUCAUAGACAGAGACGCGCAAA------GCCAGACA ---((((((((..((((....))))((((((((..((((.(((....)))......(((((((....)))))))..)))).)))....))))).....))))))))------........ ( -29.80) >consensus ___UUUGCGUGAAUUAUGCAAAUGAGUUUGGCCCUGGCCAGUUGAAUUACGCUCAAUGGCAAUUGCAAUUGCAAGAGGUUUAGCAGCGCAGACGGAGACGACCAAAGCCAAAGCCAAACA ...((((((((...))))))))...(((((((.(((((..((((((((..(((....)))..((((....))))..)))))))).)).))).((....))............))))))). (-21.43 = -23.20 + 1.77)


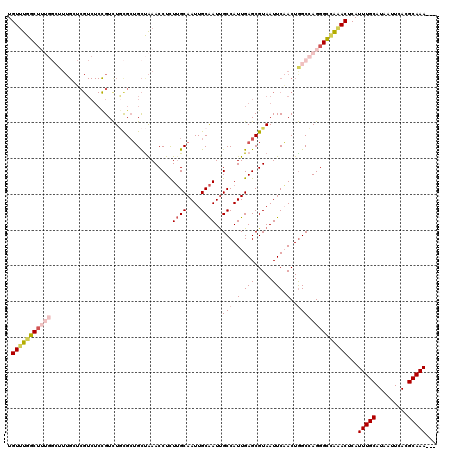
| Location | 20,618,248 – 20,618,365 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -19.58 |
| Energy contribution | -21.54 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20618248 117 - 23771897 UGUUUGGCUUUGGCUUUGGUCGUCUCCGUUUGCGCUGCUAAACCUCUUGCAAUUGCAAUCGCCAUUGAGCGUAAUUCAACUAGCCAGGGCCAAACUCAUUUGCAUAAUUCACGCAAA--- .((((((((((((((((((.((.(((((.(((((.(((..........)))..))))).)).....)))))....))))..))))))))))))))...(((((.........)))))--- ( -37.40) >DroSec_CAF1 62987 117 - 1 UGUUUGGCUUUGGCUUUGGUCGUCUCCGUCUGCGCUGCUAAACCUCUUGCAAUUGCAAUUGCCAUUGAGCGUAAUUCAACUGGCCAGGGCCGAACUCAUUUGCAUAAUUCACGCAAA--- .((((((((((((((..((......))...((((((.(.....(..((((....))))..).....)))))))........))))))))))))))...(((((.........)))))--- ( -36.60) >DroAna_CAF1 84677 102 - 1 UGUUUGGCU------------------GUCUCUGCGGUUGAACCUUUUGCAAUUGCAAUUGCCAUUGAGCGUAAUUCCACUGGCGAGGGUAAGACUCAUUUGCAUAAUUCACGCAAAGUC .(((..(((------------------((....)))))..))).((((((...(((((((((((.((..........)).))))))(((.....)))..)))))........)))))).. ( -26.20) >DroSim_CAF1 57892 117 - 1 UGUUUGGCUUUGGCUUUGCUCGUCUCCGUCUGCGCUGCUAAACCUCUUGCAAUUGCAAUUGCCAUUGAGCGUAAUUCAACUGGCCAGGGCCGAACUCAUUUGCAUAAUUCACGCAAA--- .((((((((((((((.((((((....((..((((.(((..........)))..))))..))....))))))..........))))))))))))))...(((((.........)))))--- ( -36.90) >DroEre_CAF1 55572 111 - 1 UGUCUGGC------UUUGCGCGUCUCUGUCUAUGAUGCCAAACCUUUGGCAAUUGCGAUUGCCAUUGAGCGUAAUUCUACUGGCCAGGGCUAAACUCAUUUGCAUAAUUCACGCAAA--- ((((.((.------((((.(((((.........)))))))))))...)))).(((((..(((...((((.(((....)))((((....))))..))))...))).......))))).--- ( -29.40) >consensus UGUUUGGCUUUGGCUUUGCUCGUCUCCGUCUGCGCUGCUAAACCUCUUGCAAUUGCAAUUGCCAUUGAGCGUAAUUCAACUGGCCAGGGCCAAACUCAUUUGCAUAAUUCACGCAAA___ .(((((((((((..................................((((....))))..(((((((((.....))))).)))))))))))))))...(((((.........)))))... (-19.58 = -21.54 + 1.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:09 2006