| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
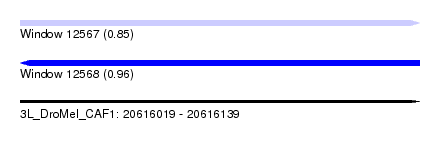
| Location | 20,616,019 – 20,616,139 |
| Length | 120 |
| Max. P | 0.958499 |

| Location | 20,616,019 – 20,616,139 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
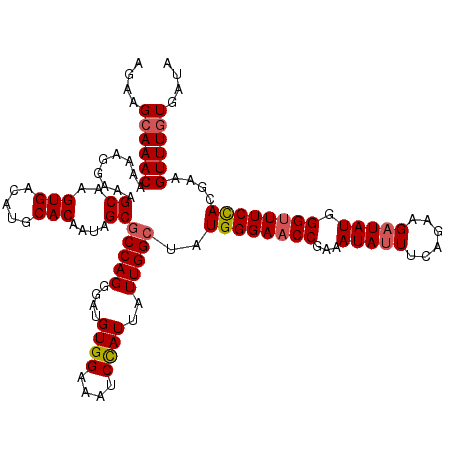
>3L_DroMel_CAF1 20616019 120 + 23771897 UAUCACAAACUUCGUGGAAACCCAUAUCUUCUGAAAUAUUUCGGUUCCCAUACCCAAUAAUGGAUUUCCACAUCCCUGGCGCUAUUGUGCAUGUCACUUGCUUUCCUUUUGUUUGCUUCU .....(((((...(((((((.((((.......(((....)))(((......))).....)))).))))))).......((((....))))....................)))))..... ( -25.80) >DroSec_CAF1 60821 120 + 1 UAUCACAAACUUCGUGGAAGCCCAUAUCUUCUGAAAUAUUUCGGUUCCCAUAGCCAAUAAUGGAUUUCCACAUCCCUGGCGCUAUUGUGCAUGUCACUUGCUUUCCUUUUGUUUGCUUCU .....(((((...((((((..((((.......(((....)))((((.....))))....))))..)))))).......((((....))))....................)))))..... ( -26.40) >DroSim_CAF1 55702 120 + 1 UAUCACAAACUUCGUGGAAACCCAUAUCUUCUGAAGUAUUUCGGUUCCCAUAGCCAAUAAUGGAUUUCCACAUCCCUGGCGCUAUUGUGCAUGUCACUUGCUUUCCUUUUGUUUGCUUCU .....(((((...(((((((.((((.((....))........((((.....))))....)))).))))))).......((((....))))....................)))))..... ( -26.30) >DroEre_CAF1 53263 110 + 1 AACUAUAAACUUCGUAGAAACCCAUA----------UAUUUCGGUUCCCAUAGCCAAUAAUAGAUUUCCACCUCCCUGGCGCUAUUGUGCAUGUCACUUGCUUUCCUUUUGUUUGCUUCU .............(((((........----------......((((.....)))).(((((((....(((......)))..)))))))(((.......)))..........))))).... ( -13.70) >consensus UAUCACAAACUUCGUGGAAACCCAUAUCUUCUGAAAUAUUUCGGUUCCCAUAGCCAAUAAUGGAUUUCCACAUCCCUGGCGCUAUUGUGCAUGUCACUUGCUUUCCUUUUGUUUGCUUCU .....(((((...(((((((.((((.((....))........((((.....))))....)))).))))))).......((((....))))....................)))))..... (-20.23 = -20.73 + 0.50)



| Location | 20,616,019 – 20,616,139 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -25.80 |
| Energy contribution | -26.55 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20616019 120 - 23771897 AGAAGCAAACAAAAGGAAAGCAAGUGACAUGCACAAUAGCGCCAGGGAUGUGGAAAUCCAUUAUUGGGUAUGGGAACCGAAAUAUUUCAGAAGAUAUGGGUUUCCACGAAGUUUGUGAUA ....((((((....((...((..(((.....)))....)).)).....(((((((((((((.(((.(((......)))(((....)))....))))))))))))))))..)))))).... ( -32.80) >DroSec_CAF1 60821 120 - 1 AGAAGCAAACAAAAGGAAAGCAAGUGACAUGCACAAUAGCGCCAGGGAUGUGGAAAUCCAUUAUUGGCUAUGGGAACCGAAAUAUUUCAGAAGAUAUGGGCUUCCACGAAGUUUGUGAUA ....((((((....((...((..(((.....)))....)).)).....(((((((..((((.(((..((..((...))(((....)))))..)))))))..)))))))..)))))).... ( -29.20) >DroSim_CAF1 55702 120 - 1 AGAAGCAAACAAAAGGAAAGCAAGUGACAUGCACAAUAGCGCCAGGGAUGUGGAAAUCCAUUAUUGGCUAUGGGAACCGAAAUACUUCAGAAGAUAUGGGUUUCCACGAAGUUUGUGAUA ....((((((....((...((..(((.....)))....)).)).....(((((((((((((.(((..((..((...........))..))..))))))))))))))))..)))))).... ( -33.00) >DroEre_CAF1 53263 110 - 1 AGAAGCAAACAAAAGGAAAGCAAGUGACAUGCACAAUAGCGCCAGGGAGGUGGAAAUCUAUUAUUGGCUAUGGGAACCGAAAUA----------UAUGGGUUUCUACGAAGUUUAUAGUU ..((((.............((..(((.....)))....))(((((..((((....))))....)))))..((((((((.(....----------..).))))))))....))))...... ( -23.30) >consensus AGAAGCAAACAAAAGGAAAGCAAGUGACAUGCACAAUAGCGCCAGGGAUGUGGAAAUCCAUUAUUGGCUAUGGGAACCGAAAUAUUUCAGAAGAUAUGGGUUUCCACGAAGUUUGUGAUA ....((((((.........((..(((.....)))....))(((((....((((....))))..)))))..((((((((...(((((......))))).))))))))....)))))).... (-25.80 = -26.55 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:03 2006