
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,615,538 – 20,615,750 |
| Length | 212 |
| Max. P | 0.642903 |

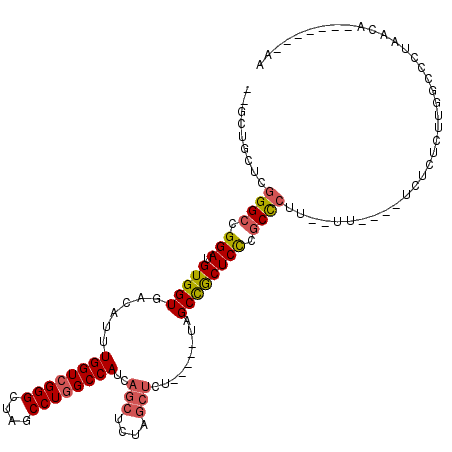
| Location | 20,615,538 – 20,615,639 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -29.92 |
| Energy contribution | -30.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20615538 101 + 23771897 --GCUGCUCGGGCCGGAUGUGGUGACAUUUGGUCGGGCUAGCCUGGCCAUCAGCUCUAGCUCU------AGCCGCUCCCGCCCUUUUUU----UCUCUCUUGGCCCUAACA-------AA --.......((((.(((.(((((......((((((((....))))))))..(((....)))..------.)))))))).))))......----..................-------.. ( -35.50) >DroSec_CAF1 60315 105 + 1 --GCUGCUCGGGCCGGAUGUGGUGACAUUUGGUCGGGCUAGCCUGGCCAUCAGCUCUAGCUCUAGCCCUAGCCGCUCCCGCCCUU--UU----UCUCUCUUGGCCCUAACA-------AA --.......((((.(((.((((((((.....)))(((((((.((((.........))))..)))))))..)))))))).))))..--..----..................-------.. ( -36.90) >DroAna_CAF1 82242 111 + 1 GCGCGACUCAGGCCGGAUGUGGUGACAUUUGGUCGGGCUGGCCUAGCCAGCUAUUCG---------GAUGGCCACUCUCUCUAUUGGUUGCAGUCCCUUAUGUCCCUAACACUAUGUAUG ..(((((.(((...(((.(((((..(((((((...(((((((...)))))))..)))---------)))))))))..)))...))))))))............................. ( -35.70) >DroSim_CAF1 55195 106 + 1 --GCUGCUCGGGCCGGAUGUGGUGACAUUUGGUCGGGCUAGCCUGGCCAUCAGCUCUAGCUCUAGCCCUAGCCGCUCCCGCCCUUU-UU----UCUCUCUUGGCCCUAACA-------AA --.......((((.(((.((((((((.....)))(((((((.((((.........))))..)))))))..)))))))).))))...-..----..................-------.. ( -36.90) >DroEre_CAF1 52844 99 + 1 --GCUGCUCGGGCCGGAUGUGGUGACAUUUGGUCGGGCUGGCCUGGCCAUCAGCUCUAGCUCU------AGCUGCUCCCGCCCUU--UU----UCUCUCUGGGCCCUAACA-------AA --((.(((.((((..((.(((....))).((((((((....)))))))).....))..)))).------))).))....((((..--..----.......)))).......-------.. ( -38.00) >consensus __GCUGCUCGGGCCGGAUGUGGUGACAUUUGGUCGGGCUAGCCUGGCCAUCAGCUCUAGCUCU_____UAGCCGCUCCCGCCCUU__UU____UCUCUCUUGGCCCUAACA_______AA .........((((.(((.(((((......((((((((....))))))))..(((....))).........)))))))).))))..................................... (-29.92 = -30.48 + 0.56)

| Location | 20,615,639 – 20,615,750 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20615639 111 + 23771897 UAUUGGUUGCAAUAAAAAGCUAAUAAAGAAUAUGGAAAG-GCAAAUGCCUGGCUGACAAGUGCAUGGAG-CCCAGGGAUGGAAAAUAGUUUUUCCGUGUAAAGAGCUAUU-------UGA ((((((((.........))))))))....((((((((((-((..((.(((((((..((......)).))-.))))).))........))))))))))))...........-------... ( -26.50) >DroSec_CAF1 60420 118 + 1 UAUUGGUUGCAAUAAAAAGCUAAUAAAGAACAUGGAAAG-ACAAAUGCCUGGCUGACAAGUGCAUGGAG-GCCAGGGAUGGAAAAUAGUUUUUCCUCGUAAAGAGCCAUUUAACAUUUGG ((((((((.........))))))))..............-.(((((((((((((..((......))..)-))))))((.((((((....))))))))................)))))). ( -32.00) >DroAna_CAF1 82353 91 + 1 UAUUGUUGGUAAUAAAAAGCUAAUAAAGAAAAUGGUUAG-ACAAAUGUCUGACUGACAAGUGCUUGGUUUGCCAGGGCUGGAAUACAAUUAG---------------------------- ..((((((((........)))))))).......((((((-((....))))))))....(((.(((((....)))))))).............---------------------------- ( -22.70) >DroSim_CAF1 55301 119 + 1 UAUUGGUUGCAAUAAAAAGCUAAUAAAGAAUAUGGAAAGAACAAAUGCCUGGCUGACAAGUGCAUGGUG-CCCAGGGAUGGAAAAUAGUUUUUCCUCGUAAAGAGCCAUUUAACAUUUGG ((((((((.........))))))))................((((((((((((...((......))..)-.)))))((.((((((....))))))))................)))))). ( -24.70) >DroEre_CAF1 52943 115 + 1 UAUUGGUUGCAAUAAAAAGCUAAUAAAGAAUAUGGAAAG-GCAAAUGCU--GCUGACAAGUGCAUGGAG-CCCAGGGAUGGGUAAUAGUU-UUCCUAGUACAGAGGCAUUCUAAUUUUGG ((((((((.........)))))))).(((((.(((((.(-((....)))--(((.....((((..((((-((((....))))).......-.)))..))))...))).)))))))))).. ( -26.20) >consensus UAUUGGUUGCAAUAAAAAGCUAAUAAAGAAUAUGGAAAG_ACAAAUGCCUGGCUGACAAGUGCAUGGAG_CCCAGGGAUGGAAAAUAGUUUUUCCUCGUAAAGAGCCAUU__A__UUUGG ((((((((.........))))))))........((((((.((..((((...(((....)))))))......(((....)))......))))))))......................... (-10.72 = -11.64 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:01 2006