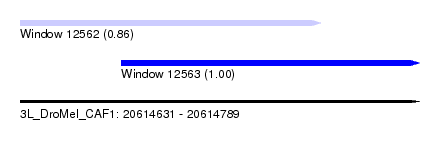
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,614,631 – 20,614,789 |
| Length | 158 |
| Max. P | 0.996834 |

| Location | 20,614,631 – 20,614,750 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -27.46 |
| Energy contribution | -26.98 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20614631 119 + 23771897 ACAGCGGCGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACA ...((.((........((((.(..((((....((.....))....))))..)..))))....(((((((((((((((.....))))).))))))))))-)).))................ ( -30.80) >DroSec_CAF1 59400 119 + 1 GCAGCGGCGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACA ...((.((........((((.(..((((....((.....))....))))..)..))))....(((((((((((((((.....))))).))))))))))-)).))................ ( -30.80) >DroAna_CAF1 81372 120 + 1 GCGGCUAUGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAGCAGAUUUGCUGUCAAGGUUGGAAUAAUUUUCCGAGUCUUGGCGGGGCUGCAAAUUAAUCAAGCACA ...(((..(((....)))..(((((((((...(((........))).......((((.....(((((((((((((((.....)))))).)))))))))..))))))))))))).)))... ( -31.70) >DroSim_CAF1 54283 119 + 1 GCAGCGGCGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACA ...((.((........((((.(..((((....((.....))....))))..)..))))....(((((((((((((((.....))))).))))))))))-)).))................ ( -30.80) >DroEre_CAF1 51920 119 + 1 GCGGCGGCGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAACAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGACGG-GCGGCAAAUUAAUCAGGCACA ((.((((((((.....((((((.......)).(((........)))........))))))))))(((((((((((((.....))))).))))))))..-)).))................ ( -31.80) >consensus GCAGCGGCGAAACUUUUCUGGGUUAAUUUUCAUGUUUUUCAAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG_GCGGCAAAUUAAUCAAGCACA ...((...(((....)))..(((((((((.(.(((........)))................(((((((((((((((.....))))).))))))))))....).)))))))))..))... (-27.46 = -26.98 + -0.48)

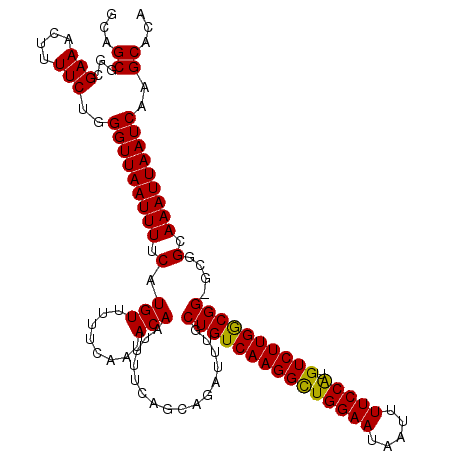
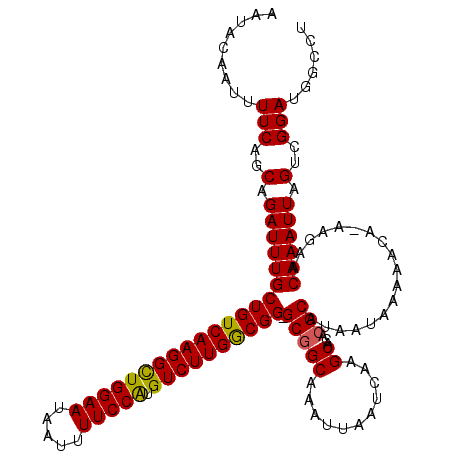
| Location | 20,614,671 – 20,614,789 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20614671 118 + 23771897 AAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACACGCAUAAUAAAAACA-AAGAACAAAUUAGUCGGAUGCCCU .........(((.((.(((((((((((((((((((((.....))))).))))))))))-(((..................)))............-.....)))))).)).)))...... ( -29.37) >DroSec_CAF1 59440 118 + 1 AAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACACGCAUAAUAAAAACA-AAGAACAAAUUAGUCGGAUGGCCU .........(((.((.(((((((((((((((((((((.....))))).))))))))))-(((..................)))............-.....)))))).)).)))...... ( -29.37) >DroAna_CAF1 81412 120 + 1 AAUACAAUUUUCAGCAGAUUUGCUGUCAAGGUUGGAAUAAUUUUCCGAGUCUUGGCGGGGCUGCAAAUUAAUCAAGCACACGCAUAACAAAAACAAAAGUACAAAUUGGAGGGAUGACCU ....((((((...((((.....(((((((((((((((.....)))))).)))))))))..))))...........((....))...................)))))).(((.....))) ( -29.70) >DroSim_CAF1 54323 118 + 1 AAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG-GCGGCAAAUUAAUCAAGCACACGCAUAAUAAAAACA-AAGAACAAAUUAGUCGGAUGGCCU .........(((.((.(((((((((((((((((((((.....))))).))))))))))-(((..................)))............-.....)))))).)).)))...... ( -29.37) >DroEre_CAF1 51960 116 + 1 AAUACAAUUUUCAACAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGACGG-GCGGCAAAUUAAUCAGGCACACGCAUAA--AAAACA-AAGAACAAAUUAGACGGAUGGCCU ................(((((((((((((((((((((.....))))).))))))))..-..))))))))....((((...((..(((--......-.........)))..))....)))) ( -29.86) >consensus AAUACAAUUUUCAGCAGAUUUGCUGUCAAGGCUGGAAUAAUUUUCCAUGUCUUGGCGG_GCGGCAAAUUAAUCAAGCACACGCAUAAUAAAAACA_AAGAACAAAUUAGUCGGAUGGCCU .........(((..(.(((((((((((((((((((((.....))))).)))))))))).(((((...........))...)))..................)))))).)..)))...... (-25.78 = -25.50 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:58 2006