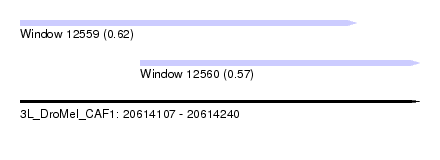
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,614,107 – 20,614,240 |
| Length | 133 |
| Max. P | 0.621002 |

| Location | 20,614,107 – 20,614,219 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -26.39 |
| Energy contribution | -27.07 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
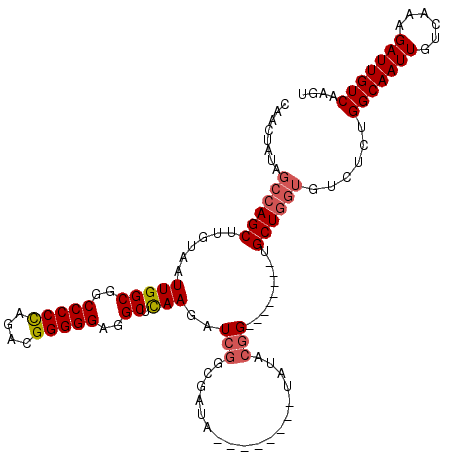
>3L_DroMel_CAF1 20614107 112 + 23771897 CAACUAUAGCCAGCUUGUAAUUGGCGGCCCCCAGACGGGGGAGGCUCAAGAUCGGCGAUAUGUACAUAUAUACGG--------UGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGU ........((((((......(((((..(((((....)))))..)).))).((((...((((....))))...)))--------))))))).....((((((((......))))))))... ( -38.10) >DroSec_CAF1 58880 112 + 1 CAACUAUAGCCAGCUUGUAAUUGGCGGCCCCUAGACGGGGGAGGCUCAAGAUCGGCGAUAUGUACAUAUAUACGG--------UGCUGGUGUCCCUGGCAAUUGUCAAAGAUUGUCAAGU ........((((((......(((((..(((((....)))))..)).))).((((...((((....))))...)))--------))))))).....((((((((......))))))))... ( -35.70) >DroAna_CAF1 80891 103 + 1 CAACUAUAGCCAGCUUGUAAUUGGCGUCCCCCAGACGGGGGAAGCUUAAAAUCC-A------------GAUAAGGGGGCGCACUGCUUA----GGGGGCAAUUGUCAAAGAUUGUCAAGU ........(((((.......))))).((((((....)))))).((((.....((-.------------..((((.((.....)).))))----.))(((((((......))))))))))) ( -32.90) >DroSim_CAF1 53782 104 + 1 CAACUAUAGCCAGCUUGUAAUUGGCGGCCCCCAGACGGGGGAGGCUCAAGAUCGGCGAUA--------UAUACAG--------UGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGU ........(((((((((((.(((((..(((((....)))))..)).))).(((...))).--------..)))))--------.)))))).....((((((((......))))))))... ( -35.90) >DroEre_CAF1 51455 104 + 1 CAACUAUAGCCAGCUUGUAAUUGGCGGCCCCCAGACGGGGGAGGCUCAAGAUCGGAGAUA--------UAUACGG--------AGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGU ........((((((((....(((((..(((((....)))))..)).)))...((......--------....)))--------))))))).....((((((((......))))))))... ( -36.10) >consensus CAACUAUAGCCAGCUUGUAAUUGGCGGCCCCCAGACGGGGGAGGCUCAAGAUCGGCGAUA________UAUACGG________UGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGU ........((((((......(((((..(((((....)))))..)).)))..(((..................))).........))))))......(((((((......))))))).... (-26.39 = -27.07 + 0.68)


| Location | 20,614,147 – 20,614,240 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20614147 93 + 23771897 GAGGCUCAAGAUCGGCGAUAUGUACAUAUAUACGGUGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGUUUUUGGCCUCUUGGUCGUGUG- (((((.(((((((((((...((((......)))).))))))).....((((((((......))))))))....)))))))))...........- ( -28.20) >DroSec_CAF1 58920 93 + 1 GAGGCUCAAGAUCGGCGAUAUGUACAUAUAUACGGUGCUGGUGUCCCUGGCAAUUGUCAAAGAUUGUCAAGUUUUUGGCCUCUUGGUCGUGUG- (((((.(((((.(((.(((((((((.........))))..)))))))((((((((......)))))))).).))))))))))...........- ( -28.30) >DroSim_CAF1 53822 85 + 1 GAGGCUCAAGAUCGGCGAUA--------UAUACAGUGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGUUUUUGGCCUCUUGGUCGUGUG- ((((((.(((((.((((((.--------...(((((((..(.....)..)).))))).....))))))..))))).))))))...........- ( -26.60) >DroEre_CAF1 51495 86 + 1 GAGGCUCAAGAUCGGAGAUA--------UAUACGGAGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGUUUUUGGCCUCUCGGUCGUGUGC ...((.((.((((((((...--------....((((((....).)))))......((((((((((....))))))))))))).))))).)).)) ( -27.40) >consensus GAGGCUCAAGAUCGGCGAUA________UAUACGGUGCUGGUGUCUCUGGCAAUUGUCAAAGAUUGUCAAGUUUUUGGCCUCUUGGUCGUGUG_ (((((.((((((((((.(((((....))))).....)))))).....((((((((......))))))))....)))))))))............ (-22.20 = -22.70 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:55 2006