
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,613,831 – 20,614,056 |
| Length | 225 |
| Max. P | 0.954982 |

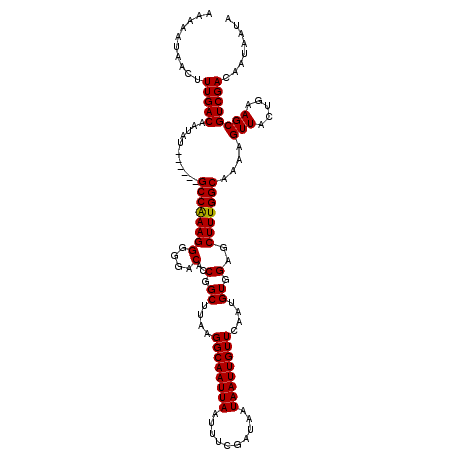
| Location | 20,613,831 – 20,613,945 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.56 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20613831 114 + 23771897 AAAAAUAACUUUGACAAUAU------GCCAAAGGGGGACACCGGCUUAAGGCAAUUAAUUUCGAUAAUAAUUGUUCAAUGUGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((....(------((((((((....).(((.(....((((((((..........))))))))....))))..))))))))...(((.....))))))))........ ( -24.40) >DroSec_CAF1 58604 114 + 1 AAAAAUAACUUUGACAAUAU------GCCAAAGGGGGACACCGGCUUAAGGCAAUUAAUUUCGAUAAUAAUUGUUCAAUGUGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((....(------((((((((....).(((.(....((((((((..........))))))))....))))..))))))))...(((.....))))))))........ ( -24.40) >DroAna_CAF1 80605 120 + 1 AAAAAUAACUUUGACAAUAUCAGUAAGCCGAAGGGGGACAUCGGCUUAAGGCAAUUAAUUUCUAUAAUAAUUGUUCAAUGUAGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((....((..((((((((..(....).)))))))).((((((((..........)))))))).......))((((..(.........)..)))))))))........ ( -28.40) >DroSim_CAF1 53506 114 + 1 AAAAAUAACUUUGACAAUAU------GCCAAAGGGGGACACCGGCUUAAGGCAAUUAAUUUCGAUAAUAAUUGUUCAAUGUGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((....(------((((((((....).(((.(....((((((((..........))))))))....))))..))))))))...(((.....))))))))........ ( -24.40) >DroEre_CAF1 51144 114 + 1 AAAAAUAACUUUGACAAUAU------GCCAAAGGGGGACACCGGCUUAAGGCAAUUAAUUUCGAUAAUAAUUGUUCAAUGUGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((....(------((((((((....).(((.(....((((((((..........))))))))....))))..))))))))...(((.....))))))))........ ( -24.40) >consensus AAAAAUAACUUUGACAAUAU______GCCAAAGGGGGACACCGGCUUAAGGCAAUUAAUUUCGAUAAUAAUUGUUCAAUGUGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUA ..........(((((...........((((((((....)..(.((....((((((((..........))))))))....)).)..)))))))....(((.....))))))))........ (-22.98 = -22.82 + -0.16)


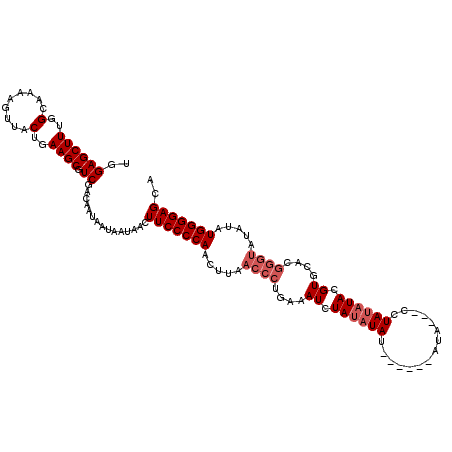
| Location | 20,613,905 – 20,614,016 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -20.16 |
| Energy contribution | -21.36 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20613905 111 + 23771897 UGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCA .(..((((..(.........)..))))..)...............(((((((.....((((((.....((((((.------...---..))))))....)).)))).....))))))).. ( -26.50) >DroSec_CAF1 58678 111 + 1 UGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUUUACGUGCACGGGUAUAUAUGGGGAGCA .(..((((..(.........)..))))..)...............((((((..(........)........((((------(((---(((............)))))))))))))))).. ( -24.50) >DroAna_CAF1 80685 106 + 1 UAGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUAGCCUCUAGAUACGUG--------UAUAUGGGGAGCA ..((((((..(.........)..)))).))...............(((((((.(........).....((((((.------.((.........))..))--------))))))))))).. ( -18.50) >DroSim_CAF1 53580 111 + 1 UGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCA .(..((((..(.........)..))))..)...............(((((((.....((((((.....((((((.------...---..))))))....)).)))).....))))))).. ( -26.50) >DroEre_CAF1 51218 117 + 1 UGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAUACACGUAUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCA .(..((((..(.........)..))))..)...............(((((((.....((((.(....)........((((((((---....))))))))...)))).....))))))).. ( -30.20) >consensus UGGAGCUUUGGCAAAAGUUACUGAAGCGUCGACAAUAAUAAUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU______AUA___CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCA ..((((((..(.........)..)))).))...............(((((((.....((((....((.((((((...............)))))).))....)))).....))))))).. (-20.16 = -21.36 + 1.20)

| Location | 20,613,945 – 20,614,056 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20613945 111 + 23771897 AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....(((((((.....((((((.....((((((.------...---..))))))....)).)))).....)))))))....(((((...(((((.......)))))......))))).. ( -25.50) >DroSec_CAF1 58718 111 + 1 AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUUUACGUGCACGGGUAUAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....((((((..(........)........((((------(((---(((............))))))))))))))))....(((((...(((((.......)))))......))))).. ( -23.50) >DroAna_CAF1 80725 106 + 1 AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUAGCCUCUAGAUACGUG--------UAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....(((((((.(........).....((((((.------.((.........))..))--------)))))))))))....(((((...(((((.......)))))......))))).. ( -17.90) >DroSim_CAF1 53620 111 + 1 AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU------AUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....(((((((.....((((((.....((((((.------...---..))))))....)).)))).....)))))))....(((((...(((((.......)))))......))))).. ( -25.50) >DroEre_CAF1 51258 117 + 1 AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAUACACGUAUA---CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....(((((((.....((((.(....)........((((((((---....))))))))...)))).....)))))))....(((((...(((((.......)))))......))))).. ( -29.20) >consensus AUAACUUCCCCAACUUAACCCUGAAAUCUAUAUAU______AUA___CCUAUAUACGUGCACGGGUAUAUAUGGGGAGCAAGUCAUUAACGAGCUCUCGAACAGCUCUCGAACAAUGAAC .....(((((((.....((((....((.((((((...............)))))).))....)))).....)))))))....(((((...(((((.......)))))......))))).. (-19.72 = -20.92 + 1.20)

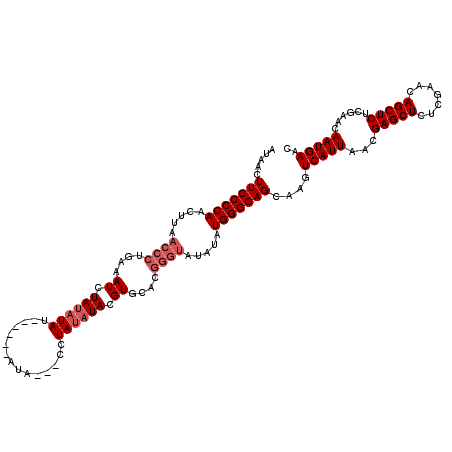

| Location | 20,613,945 – 20,614,056 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20613945 111 - 23771897 GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUAUACCCGUGCACGUAUAUAGG---UAU------AUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((.....((((......(((((((..---..)------))))))........)))).....))))).)))... ( -31.94) >DroSec_CAF1 58718 111 - 1 GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUAUACCCGUGCACGUAAAUAGG---UAU------AUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((((((((((..(........)..))---)))------))))...(((((.......))))))))).)))... ( -29.40) >DroAna_CAF1 80725 106 - 1 GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUA--------CACGUAUCUAGAGGCUAU------AUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((....--------.((..((((.((((((((------....)))).)))).))))...))))))).)))... ( -27.60) >DroSim_CAF1 53620 111 - 1 GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUAUACCCGUGCACGUAUAUAGG---UAU------AUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((.....((((......(((((((..---..)------))))))........)))).....))))).)))... ( -31.94) >DroEre_CAF1 51258 117 - 1 GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUAUACCCGUGCACGUAUAUAGG---UAUACGUGUAUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((.....(((((((((((((((....---))))))))))).....(....).)))).....))))).)))... ( -41.10) >consensus GUUCAUUGUUCGAGAGCUGUUCGAGAGCUCGUUAAUGACUUGCUCCCCAUAUAUACCCGUGCACGUAUAUAGG___UAU______AUAUAUAGAUUUCAGGGUUAAGUUGGGGAAGUUAU ..((((((..((((..((.....))..)))).))))))...((((((((.....((((.......((((((...............)))))).......)))).....))))).)))... (-25.70 = -26.90 + 1.20)

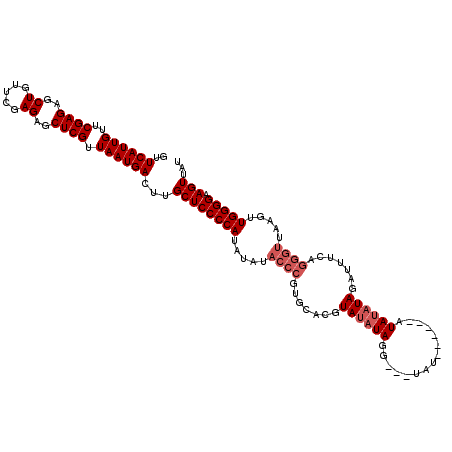
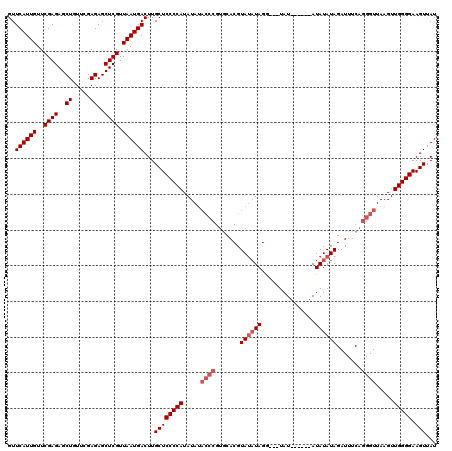
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:53 2006