| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,308,135 – 2,308,230 |
| Length | 95 |
| Max. P | 0.500000 |

| Location | 2,308,135 – 2,308,230 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.18 |
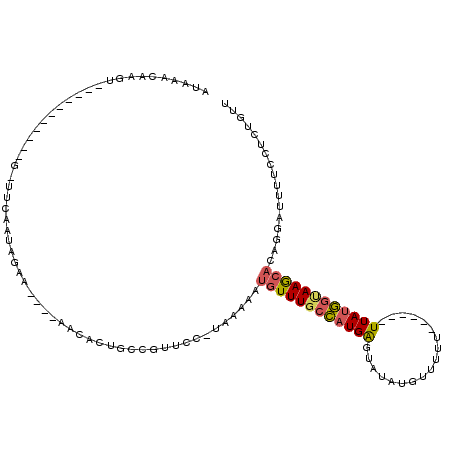
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2308135 95 + 23771897 AUAAACAAGU-----------G-CUCAAUAGAA----UACACUGCCGUUCCGUGAAUAUGUUUGCCAUGAGUAUAUGUUUUU------UUAUGGUAAGCAGAGGAUUUUUCUCUGUU ..(((((.((-----------(-((((...(((----(........)))).(..((....))..)..))))))).)))))..------........(((((((((...))))))))) ( -21.20) >DroPse_CAF1 70976 113 + 1 CUUAACAAGUCGAAAGAUACAU-UUCUAUAGAUGUGUAAGGCUGUUAUUUU-CAAGGAUCUUU--UAUGGGAAUAUAAUUUAAGUACAUUAAAAAAAAAAAAGGAUGUACCCUCGUG ..(((((.(((.....((((((-(......)))))))..))))))))....-..(((.((((.--...))))...........(((((((.............)))))))))).... ( -19.12) >DroSec_CAF1 57280 94 + 1 AUAAAAAAGU-----------G-UUCAAUAGAA----UACACUGUCGUUCC-UAAAAAUGUUUGCCAUGAGUAUAUGUUUUU------UUAUGGUAAGCAGAAGAUUUUCCUUUGUU ........((-----------(-(((....)))----)))...........-......(((((((((((((..........)------))))))))))))((((......))))... ( -20.80) >DroSim_CAF1 57252 94 + 1 AUAAACAAGU-----------G-UUCAAUAGAA----UACACUGCCGUUCC-UAAAAAUGUUUGCCAUGAGUAUAUGUUUUU------UUAUGGUAAGCACUGGAUUUUCCUCUGUU ...((((.((-----------(-(((....)))----)))........(((-......(((((((((((((..........)------))))))))))))..)))........)))) ( -21.80) >DroEre_CAF1 60605 94 + 1 AUAAACAAGU-----------UUUUCCAUAGAA----AAGACUGCCGUUUC-UA-AAAUGUUUGCCAUGAGUAUAUGUUUUU------UUAUGGCAAGCCCAGGAUUUCCGUCUGUU ...((((..(-----------...(((.(((((----(.(.....).))))-))-....((((((((((((..........)------)))))))))))...))).....)..)))) ( -21.10) >DroYak_CAF1 59980 94 + 1 AUAAACAAGU-----------U-UUCCAUAGAA----AAGACUGACGUUUC-CAAAAAUGUUUGCCAUGAGUAUAUGUUUUU------UUAUGGCAAGCACAGGAUUUCCGUCUGUU ...(((((((-----------(-((........----))))))((((..((-(.....(((((((((((((..........)------))))))))))))..)))....)))))))) ( -25.10) >consensus AUAAACAAGU___________G_UUCAAUAGAA____AACACUGCCGUUCC_UAAAAAUGUUUGCCAUGAGUAUAUGUUUUU______UUAUGGUAAGCACAGGAUUUUCCUCUGUU ..........................................................((((((((((((..................))))))))))))................. ( -9.42 = -9.54 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:50 2006