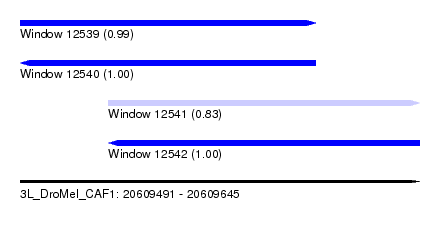
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,609,491 – 20,609,645 |
| Length | 154 |
| Max. P | 0.999341 |

| Location | 20,609,491 – 20,609,605 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609491 114 + 23771897 AAUAUGAUAUUAAAAUUGAACUUUGCCGCCG------AGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGAAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGA ....................(.((((((.((------(.(((((..((((((((((((((.........)))))..((......))....)))))))))..))))).))).)))))).). ( -34.20) >DroSec_CAF1 54350 114 + 1 AAUAUGAUAUUAAAAUUGAACUUUGCCGCCG------AGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGA ....................(.((((((.((------(.(((((..((((((((((((((.........)))))..(((....)))....)))))))))..))))).))).)))))).). ( -36.90) >DroAna_CAF1 76164 120 + 1 AAUAUGAUAUUAAAAUUGAACUUUGCCGCCGUCUGGUAGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGCCUGUAACUAAUUAAAUGCACUUGCAAAUUGCCGGCAACGA .............................(((((((((((((((..(((((((((...(((.....)))..(((....))).........)))))))))..))).)))))))))..))). ( -32.70) >DroSim_CAF1 49180 114 + 1 AAUAUGAUAUUAAAAUUGAACUUUGCCGCCG------AGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGACAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGA ....................(.((((((.((------(.(((((..((((((((((((((((.((......)).....)))))..)))...))))))))..))))).))).)))))).). ( -32.30) >DroEre_CAF1 46835 114 + 1 AAUAUGAUAUUAAAAUUGAACUUUGCCGCCG------AGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGGCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGA ....................(.((((((.((------(.(((((..((((((((((((((.........)))))...((((....)))).)))))))))..))))).))).)))))).). ( -37.50) >consensus AAUAUGAUAUUAAAAUUGAACUUUGCCGCCG______AGUUGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGA ......................(((.(((((........(((((..((((((((((((((.........)))))..(((....)))....)))))))))..))))).....))))).))) (-28.90 = -29.58 + 0.68)

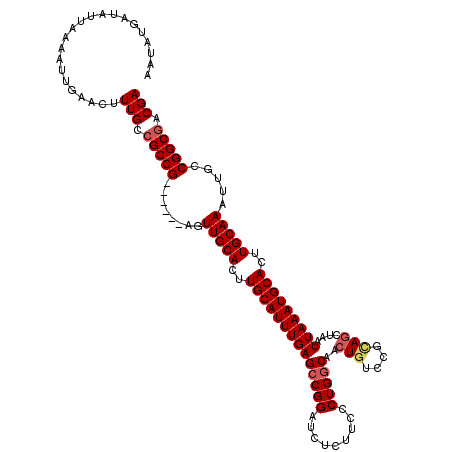
| Location | 20,609,491 – 20,609,605 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.88 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609491 114 - 23771897 UCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUUCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACU------CGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..((((((((....(((((..(((((((((((..(....)..))))(((.(((......))))))..)))))))..))))).)------)))))))........................ ( -36.40) >DroSec_CAF1 54350 114 - 1 UCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACU------CGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..((((((((....(((((..(((((((....(((((....)))))(((.(((......))))))..)))))))..))))).)------)))))))........................ ( -37.80) >DroAna_CAF1 76164 120 - 1 UCGUUGCCGGCAAUUUGCAAGUGCAUUUAAUUAGUUACAGGCAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACUACCAGACGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..(((((((.....(((((..(((((((....((((...(((....)))..(((.....))))))).)))))))..)))))........)))))))........................ ( -33.52) >DroSim_CAF1 49180 114 - 1 UCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGUCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACU------CGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..((((((((....(((((..(((((((....((((...(((....)))..(((.....))))))).)))))))..))))).)------)))))))........................ ( -36.40) >DroEre_CAF1 46835 114 - 1 UCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGCCAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACU------CGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..((((((((....(((((..(((((((....((((..(((.....)))..(((.....))))))).)))))))..))))).)------)))))))........................ ( -37.80) >consensus UCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCAACU______CGGCGGCAAAGUUCAAUUUUAAUAUCAUAUU ..(((((((.....(((((..(((((((....(((((....)))))(((.(((......))))))..)))))))..)))))........)))))))........................ (-32.96 = -32.88 + -0.08)

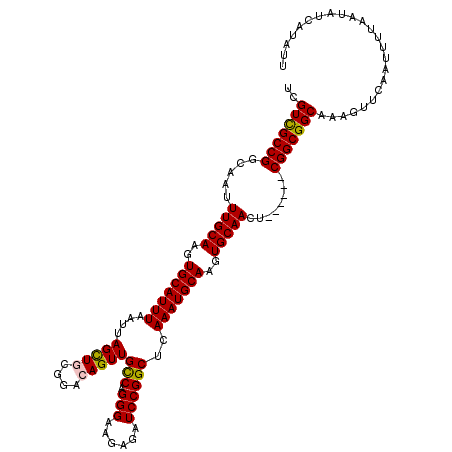
| Location | 20,609,525 – 20,609,645 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.04 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609525 120 + 23771897 UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGAAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGAAUCAUUUAUUGAGUUAUUCAAAUGAAAUGCCAGCUAUGUG ((((..((((((((((((((.........)))))..((......))....)))))))))..))))....((.((((.....(((.....)))....(((....))).)))).))...... ( -29.70) >DroSec_CAF1 54384 120 + 1 UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGAAUCAUUUAUUGAGUUAUUCAAAUGAAAUGCCAGCUAUGUG ((((..((((((((((((((.........)))))..(((....)))....)))))))))..))))....((.((((.....(((.....)))....(((....))).)))).))...... ( -32.40) >DroAna_CAF1 76204 120 + 1 UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGCCUGUAACUAAUUAAAUGCACUUGCAAAUUGCCGGCAACGAAUCACUUAUUGAGUUAUUCAAAUGAAAUGCCAGCAAUGCG ((((..(((((((((...(((.....)))..(((....))).........)))))))))..)))).(((((.((((..((((.((((...)))).))))........)))).)))))... ( -33.20) >DroSim_CAF1 49214 120 + 1 UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGACAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGAAUCAUUUAUUGAGUUAUUCAAAUGAAAUGCCAGCUAUGUG ((((..((((((((((((((((.((......)).....)))))..)))...))))))))..))))....((.((((.....(((.....)))....(((....))).)))).))...... ( -27.80) >DroEre_CAF1 46869 120 + 1 UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGGCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGAAUCAUUUAUUGAGUUAUUCAAAUGAAAUGCCAGCUAUGUG ((((..((((((((((((((.........)))))...((((....)))).)))))))))..))))....((.((((.....(((.....)))....(((....))).)))).))...... ( -33.00) >consensus UGCACUUGCAUUUGAGCCGGAUCUCUUCCCUGGCAACUGUCCGCAGCUAAUUAAAUGCACUUGCAAAUUGCCGGCGACGAAUCAUUUAUUGAGUUAUUCAAAUGAAAUGCCAGCUAUGUG ((((..((((((((((((((.........)))))..(((....)))....)))))))))..))))....((.((((.....(((.....)))....(((....))).)))).))...... (-28.72 = -29.04 + 0.32)


| Location | 20,609,525 – 20,609,645 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -32.94 |
| Energy contribution | -32.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609525 120 - 23771897 CACAUAGCUGGCAUUUCAUUUGAAUAACUCAAUAAAUGAUUCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUUCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ......((((((((.(((((((..........)))))))...)).))))))....((((..(((((((((((..(....)..))))(((.(((......))))))..)))))))..)))) ( -34.40) >DroSec_CAF1 54384 120 - 1 CACAUAGCUGGCAUUUCAUUUGAAUAACUCAAUAAAUGAUUCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ......((((((((.(((((((..........)))))))...)).))))))....((((..(((((((....(((((....)))))(((.(((......))))))..)))))))..)))) ( -35.80) >DroAna_CAF1 76204 120 - 1 CGCAUUGCUGGCAUUUCAUUUGAAUAACUCAAUAAGUGAUUCGUUGCCGGCAAUUUGCAAGUGCAUUUAAUUAGUUACAGGCAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ...((((((((((..((((((((.....))))...)))).....)))))))))).((((..(((((((....((((...(((....)))..(((.....))))))).)))))))..)))) ( -38.70) >DroSim_CAF1 49214 120 - 1 CACAUAGCUGGCAUUUCAUUUGAAUAACUCAAUAAAUGAUUCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGUCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ......((((((((.(((((((..........)))))))...)).))))))....((((..(((((((....((((...(((....)))..(((.....))))))).)))))))..)))) ( -34.40) >DroEre_CAF1 46869 120 - 1 CACAUAGCUGGCAUUUCAUUUGAAUAACUCAAUAAAUGAUUCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGCCAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ......((((((((.(((((((..........)))))))...)).))))))....((((..(((((((....((((..(((.....)))..(((.....))))))).)))))))..)))) ( -35.80) >consensus CACAUAGCUGGCAUUUCAUUUGAAUAACUCAAUAAAUGAUUCGUCGCCGGCAAUUUGCAAGUGCAUUUAAUUAGCUGCGGACAGUUGCCAGGGAAGAGAUCCGGCUCAAAUGCAAGUGCA ......((((((((.((((((((.....))))...))))...)).))))))....((((..(((((((....(((((....)))))(((.(((......))))))..)))))))..)))) (-32.94 = -32.86 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:38 2006