
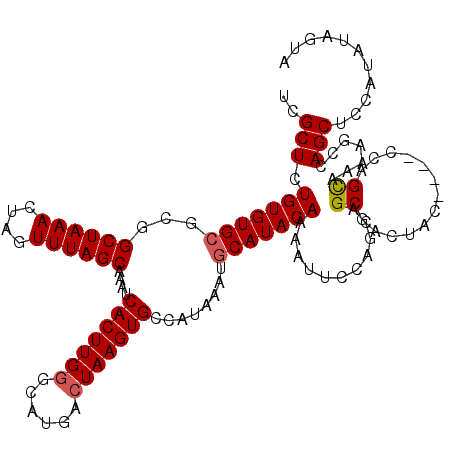
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,609,303 – 20,609,459 |
| Length | 156 |
| Max. P | 0.998722 |

| Location | 20,609,303 – 20,609,419 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609303 116 - 23771897 UCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGAGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAGCAGCGACUAC----CCAGCGAAAGCCAGCUCCAUAUAGUA ((((((((((((.(((((((((....))))))........((.((((......))))))....))).))))......))).)))))....----...((....))............... ( -27.60) >DroSec_CAF1 54162 116 - 1 UCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAGCAGCCACUAC----CCAGCAAAAGCCAGCUCCAUAUGGUA ..(((.((((((((..((((((....))))))....(((((((......))))))).......)))))))).......))).((((....----..(((........))).....)))). ( -29.50) >DroAna_CAF1 75986 116 - 1 UCGCUCUGUGUG--CGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAACGGCUAUUGCCAGCCAAGUAAAAGUCAGCCCGAGCUCG-- ..((((.(((..--..((((((....))))))....)))..((((.(((((..((((.......)))).............((((......)))).......)))))))))))))...-- ( -38.50) >DroSim_CAF1 48992 116 - 1 UCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAGCAGCGACUAC----CCAGCAAAAGCCAGCUCCAUAUAGUA .(((((((((((.(((((((((....))))))....(((((((......))))))).......))).))))......))).))))((((.----..(((........))).....)))). ( -29.00) >DroEre_CAF1 46639 116 - 1 UCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAGCAGCGCUUAC----CCAGCAAAAGCCAGCUCCAUAUAGUA ..(((..(((.((((.((((((....))))))....(((((((......))))))).......(((.............))))))).)))----..)))..................... ( -28.52) >consensus UCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAGCAGCGACUAC____CCAGCAAAAGCCAGCUCCAUAUAGUA ..(((.(((((((...((((((....))))))....(((((((......)))))))........)))))))...........((.............)).......)))........... (-23.70 = -23.94 + 0.24)


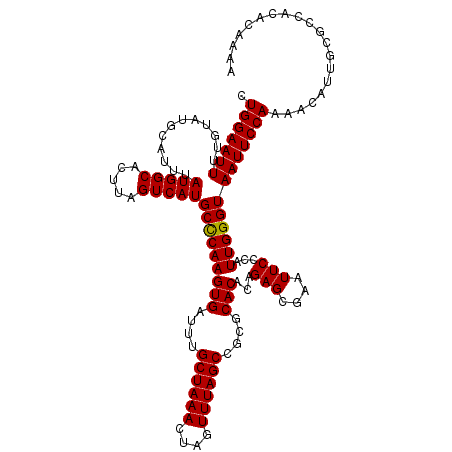
| Location | 20,609,339 – 20,609,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609339 120 + 23771897 CUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCUCAAGUGAUUUGCUAAACUAGUUUAGCCGCGCACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACAUUGCGCCACACAAAA ..(((((((...........(((((.....)))))((((..(((....((((((....))))))....)))...)))))))))))...((((((((........))))).)))....... ( -29.50) >DroSec_CAF1 54198 120 + 1 CUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCCCAAGUGAUUUGCUAAACUAGUUUAGCCGCGCACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACAUUGCGCCACACAAAA ......((((((.......((((((.....)))))).....(((....((((((....)))))).(((((........((((((((...))).)))))........)))))))))))))) ( -28.79) >DroAna_CAF1 76024 118 + 1 UUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCCCAAGUGAUUUGCUAAACUAGUUUAGCCG--CACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACCGUACACUCAACAAAA ((((((((............(((((.....)))))((((((.((....((((((....))))))((--(.......)))......))))))))))))))))................... ( -27.40) >DroSim_CAF1 49028 120 + 1 CUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCCCAAGUGAUUUGCUAAACUAGUUUAGCCGCGCACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACAUUGCGCCACACAAAA ......((((((.......((((((.....)))))).....(((....((((((....)))))).(((((........((((((((...))).)))))........)))))))))))))) ( -28.79) >DroEre_CAF1 46675 120 + 1 CUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCCCAAGUGAUUUGCUAAACUAGUUUAGCCGCGCACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACAUUGCGCCACACAAAA ......((((((.......((((((.....)))))).....(((....((((((....)))))).(((((........((((((((...))).)))))........)))))))))))))) ( -28.79) >consensus CUGGAAUUUUGUAUGCAUUUAUGGCACUUAGUCAUGCCCAAGUGAUUUGCUAAACUAGUUUAGCCGCGCACACAGAGCGAAUUCCCAUUGGGUAAUUCCAAAACAUUGCGCCACACAAAA .(((((((............(((((.....)))))(((((((((....((((((....))))))....)))...(((....)))...))))))))))))).................... (-25.66 = -25.50 + -0.16)

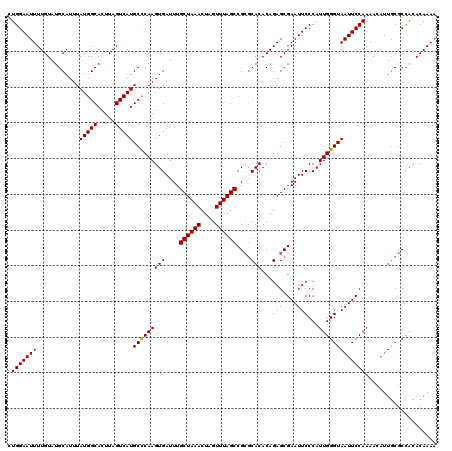
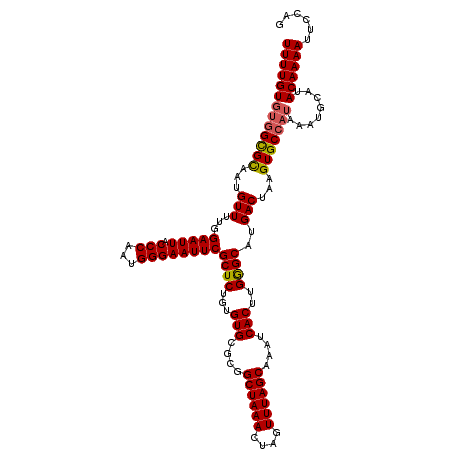
| Location | 20,609,339 – 20,609,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -32.24 |
| Energy contribution | -32.36 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20609339 120 - 23771897 UUUUGUGUGGCGCAAUGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGAGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAG .....((((((((...(((...(((((.(((...))))))))((((...(((....((((((....))))))....)))..))))..)))...))))))))................... ( -36.10) >DroSec_CAF1 54198 120 - 1 UUUUGUGUGGCGCAAUGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAG ((((((((((((((........(((((.(((...))))))))........))))).((((((....))))))....(((((((......))))))).........)))))))))...... ( -37.39) >DroAna_CAF1 76024 118 - 1 UUUUGUUGAGUGUACGGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUG--CGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAA ...................((((((((.(((...))).........((((((--((((((((....))))))....(((((((......))))))).......)))))))).)))))))) ( -33.80) >DroSim_CAF1 49028 120 - 1 UUUUGUGUGGCGCAAUGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAG ((((((((((((((........(((((.(((...))))))))........))))).((((((....))))))....(((((((......))))))).........)))))))))...... ( -37.39) >DroEre_CAF1 46675 120 - 1 UUUUGUGUGGCGCAAUGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAG ((((((((((((((........(((((.(((...))))))))........))))).((((((....))))))....(((((((......))))))).........)))))))))...... ( -37.39) >consensus UUUUGUGUGGCGCAAUGUUUUGGAAUUACCCAAUGGGAAUUCGCUCUGUGUGCGCGGCUAAACUAGUUUAGCAAAUCACUUGGGCAUGACUAAGUGCCAUAAAUGCAUACAAAAUUCCAG (((((((((((((...(((...(((((.(((...))))))))((((...(((....((((((....))))))....)))..))))..)))...)))))))........))))))...... (-32.24 = -32.36 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:34 2006