
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
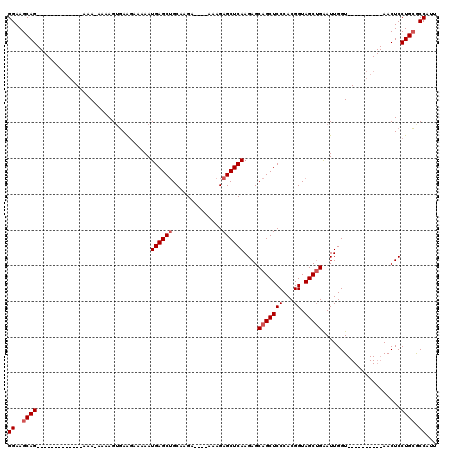
| Location | 20,606,801 – 20,606,893 |
| Length | 92 |
| Max. P | 0.629437 |

| Location | 20,606,801 – 20,606,893 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
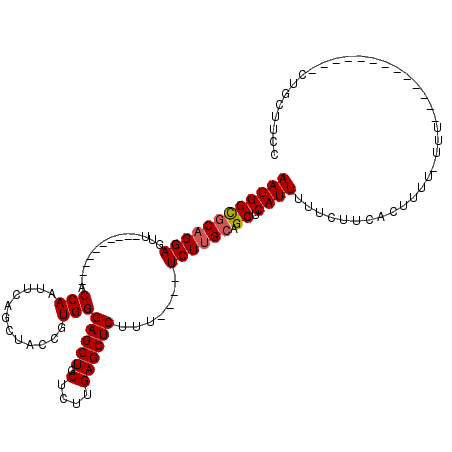
>3L_DroMel_CAF1 20606801 92 + 23771897 GGAAGCAG-------------AAA-AAAAGUGAAGAAAAAUGAGCUGCAAGA----AAAGAGCUCAAGAGCAGCUCCCACGGUAGCUGAAUUGGU----------AACUCCUGCGCCAUU ((..((((-------------...-...............((((((......----....))))))....(((((((...)).))))).......----------.....)))).))... ( -21.80) >DroSec_CAF1 51671 92 + 1 GGAAGCAG-------------AAA-AAAAGUGAAGAAAAAUGAGCUGCAAGA----AAAGAGCUCAAGAGCAGCUCCCACGGUAGCUGAAUUGGU----------AACUCCUGCGCCAUU ((..((((-------------...-...............((((((......----....))))))....(((((((...)).))))).......----------.....)))).))... ( -21.80) >DroAna_CAF1 73572 119 + 1 GGAAGCAGGGGUACUAACAAAAAA-AAAAGGCAGAAAAAAUGAGCAGCCAGAAAAAAAAGAGCUCAAGAGCAGCUCCCACGGUAGCUGGAUUGGUUUCCACAGCCAACUCCUGGGCCAUU ((...(((((((............-....(((..............)))..........((((((....).)))))....(((.(.((((......))))).))).)))))))..))... ( -29.94) >DroSim_CAF1 46466 93 + 1 GGAAGCAG-------------AAAAAAAAGUGAAGAAAAAUGAGCUGCAAGA----AAAGAGCUCAAGAGCAGCUCCCACGGUAGCUGAAUUGGU----------AACUCCUGCGCCAUU ((..((((-------------...................((((((......----....))))))....(((((((...)).))))).......----------.....)))).))... ( -21.80) >DroEre_CAF1 44200 91 + 1 GGAAGCAG-------------ACA--AAAGUGAAGAAAAAUGAGCUGCAAGA----AAAGAGCUCAAGAGCAGCUCCCACGGUAGCGGAAUUGGU----------AACUCCUGCACCAUU ((.(((..-------------((.--...)).........((((((......----....))))))......))).))..(((.(((((......----------...))).)))))... ( -19.90) >consensus GGAAGCAG_____________AAA_AAAAGUGAAGAAAAAUGAGCUGCAAGA____AAAGAGCUCAAGAGCAGCUCCCACGGUAGCUGAAUUGGU__________AACUCCUGCGCCAUU ((..((((................................((((((..............))))))....(((((((...)).)))))......................)))).))... (-17.08 = -17.68 + 0.60)


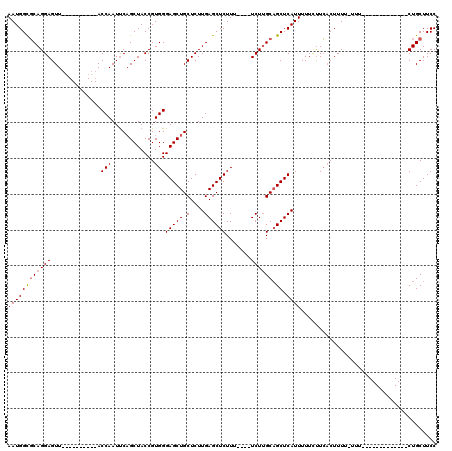
| Location | 20,606,801 – 20,606,893 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20606801 92 - 23771897 AAUGGCGCAGGAGUU----------ACCAAUUCAGCUACCGUGGGAGCUGCUCUUGAGCUCUUU----UCUUGCAGCUCAUUUUUCUUCACUUUU-UUU-------------CUGCUUCC ......(((((((..----------.(((............)))(((((((....(((.....)----))..)))))))................-)))-------------)))).... ( -22.20) >DroSec_CAF1 51671 92 - 1 AAUGGCGCAGGAGUU----------ACCAAUUCAGCUACCGUGGGAGCUGCUCUUGAGCUCUUU----UCUUGCAGCUCAUUUUUCUUCACUUUU-UUU-------------CUGCUUCC ......(((((((..----------.(((............)))(((((((....(((.....)----))..)))))))................-)))-------------)))).... ( -22.20) >DroAna_CAF1 73572 119 - 1 AAUGGCCCAGGAGUUGGCUGUGGAAACCAAUCCAGCUACCGUGGGAGCUGCUCUUGAGCUCUUUUUUUUCUGGCUGCUCAUUUUUUCUGCCUUUU-UUUUUUGUUAGUACCCCUGCUUCC ...((((((((((((((.((.(....)))..)))))).)).)))((((.(((...(((.........))).))).)))).........)))....-......(..((((....))))..) ( -29.30) >DroSim_CAF1 46466 93 - 1 AAUGGCGCAGGAGUU----------ACCAAUUCAGCUACCGUGGGAGCUGCUCUUGAGCUCUUU----UCUUGCAGCUCAUUUUUCUUCACUUUUUUUU-------------CUGCUUCC ......(((((((..----------.(((............)))(((((((....(((.....)----))..))))))).................)))-------------)))).... ( -22.20) >DroEre_CAF1 44200 91 - 1 AAUGGUGCAGGAGUU----------ACCAAUUCCGCUACCGUGGGAGCUGCUCUUGAGCUCUUU----UCUUGCAGCUCAUUUUUCUUCACUUU--UGU-------------CUGCUUCC .(((((((.(((((.----------....))))))).)))))((((((......(((((((...----....).)))))).........((...--.))-------------..)))))) ( -25.30) >consensus AAUGGCGCAGGAGUU__________ACCAAUUCAGCUACCGUGGGAGCUGCUCUUGAGCUCUUU____UCUUGCAGCUCAUUUUUCUUCACUUUU_UUU_____________CUGCUUCC ((((((((((((..............(((............)))(((((.(....)))))).......)))))).)).))))...................................... (-16.22 = -16.46 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:27 2006