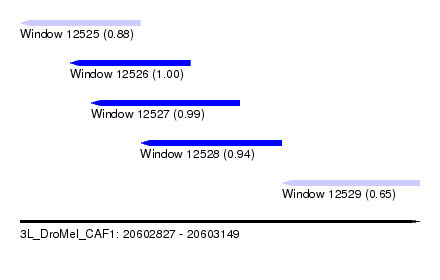
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,602,827 – 20,603,149 |
| Length | 322 |
| Max. P | 0.999868 |

| Location | 20,602,827 – 20,602,924 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20602827 97 - 23771897 UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------CCCGAUCCCCGAGUCACCCCGGACGACCACCAAGUGGACUU ...((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------........(((........)))....((((...)))).... ( -28.60) >DroSec_CAF1 47724 97 - 1 UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------CCCAAUCCCCGAGUCACUCCGGACGACCACCAAGUGGACUU ...((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------.......((.(((...))).))....((((...)))).... ( -28.70) >DroAna_CAF1 69689 115 - 1 UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAUACUUCGAUACUCGCUACCCCAUACCCAAUACCCGA----UACCCGUUGCCCGCCAAUCCGA-AC ..(((((((((((((.((((((((.......)))))))).....))))))))))))).((((....(((...................)))----.....((((.....)))).)))-). ( -22.31) >DroSim_CAF1 42513 97 - 1 UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------CCCAAUCCCCGAGUCAUCCCGGACGACCACCAAGUGGACUU ...((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------........(((........)))....((((...)))).... ( -28.60) >DroEre_CAF1 40155 96 - 1 UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------CCCAAUCCCCGUUCCACCCCGGACGACCACCAGUCCG-AUC ...((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------...................(((((........)))))-... ( -30.40) >consensus UAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC_______________________CCCAAUCCCCGAGUCACCCCGGACGACCACCAAGUGGACUU ...((((((((((((.((((((((.......)))))))).....))))))))))))...............................(((........)))................... (-22.84 = -22.88 + 0.04)


| Location | 20,602,867 – 20,602,964 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -25.88 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20602867 97 - 23771897 GAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------C ..........((....))...((((.........)))).....((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------. ( -27.80) >DroSec_CAF1 47764 97 - 1 GAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------C ..........((....))...((((.........)))).....((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------. ( -27.80) >DroAna_CAF1 69724 120 - 1 GAUAUGAAAAGAGCAACCAAGUGGCAUACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAUACUUCGAUACUCGCUACCCCAUAC ..((((.....(((.......((((.........))))....(((((((((((((.((((((((.......)))))))).....)))))))))))))...........)))....)))). ( -31.40) >DroSim_CAF1 42553 97 - 1 GAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------C ..........((....))...((((.........)))).....((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------. ( -27.80) >DroEre_CAF1 40194 97 - 1 GAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC-----------------------C ..........((....))...((((.........)))).....((((((((((((.((((((((.......)))))))).....))))))))))))-----------------------. ( -27.80) >consensus GAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCUGAACCGUUUAGUGUAC_______________________C ..........((....))...((((.........)))).....((((((((((((.((((((((.......)))))))).....))))))))))))........................ (-25.88 = -25.92 + 0.04)



| Location | 20,602,884 – 20,603,004 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.52 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20602884 120 - 23771897 AGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .((((((((.....)))))).))((((((.(((((((..((((.((.......)))))).))))).........((((...(((..(((...)))..))))))).......)))).)))) ( -33.20) >DroSec_CAF1 47781 120 - 1 AGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .((((((((.....)))))).))((((((.(((((((..((((.((.......)))))).))))).........((((...(((..(((...)))..))))))).......)))).)))) ( -33.20) >DroAna_CAF1 69764 120 - 1 AGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGAAGAUAUGAAAAGAGCAACCAAGUGGCAUACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .((((((((.....)))))).)).(((((((.(((.....)))..................((((.........))))..))))))).........((((((((.......)))))))). ( -29.50) >DroSim_CAF1 42570 120 - 1 AGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .((((((((.....)))))).))((((((.(((((((..((((.((.......)))))).))))).........((((...(((..(((...)))..))))))).......)))).)))) ( -33.20) >DroEre_CAF1 40211 120 - 1 AGACAGAGUCUGGAACUCUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .((((((((.....)))))).))((((((.(((((((..((((.((.......)))))).))))).........((((...(((..(((...)))..))))))).......)))).)))) ( -35.80) >consensus AGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAAUAUGUGCAUUAAAUGUGCAUUGGCAUUAAUCGUCGAUGCU .(((...((.((..(((((((((....(((((((....))).))))....)))))...))))..)).)).....((((...(((..(((...)))..))))))).......)))...... (-31.64 = -31.52 + -0.12)



| Location | 20,602,924 – 20,603,038 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.32 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20602924 114 - 23771897 ACAGAAUCAGAUUCAGAU------UCGGAUUCAAAGCCGGAGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAA ...((.((((((((...(------((((........)))))....))))))))....((((((....(((((((....))).))))....))))))))...((((.........)))).. ( -32.40) >DroSec_CAF1 47821 114 - 1 ACAGAAUCAGAUUCAGAU------UCGGAUUCAAAGCCGGAGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAA ...((.((((((((...(------((((........)))))....))))))))....((((((....(((((((....))).))))....))))))))...((((.........)))).. ( -32.40) >DroAna_CAF1 69804 110 - 1 ---GGAUUA-AAACAGAU------UCGAAUUCAAAGUCAGAGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGAAGAUAUGAAAAGAGCAACCAAGUGGCAUACAAAAAGCCAAA ---((....-...(((((------((...(((.......)))...))))))).....((((((....(((((........).))))....))))))))...((((.........)))).. ( -25.20) >DroSim_CAF1 42610 114 - 1 ACAGAAUCAGAUUCAGAU------UCGGACUCAAAGCCGGAGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAA ...((.((((((((...(------((((........)))))....))))))))....((((((....(((((((....))).))))....))))))))...((((.........)))).. ( -32.40) >DroEre_CAF1 40251 120 - 1 CCAGAAUCAGAUUUAGAUUCAGACUCGAAUUCAAAGCCGGAGACAGAGUCUGGAACUCUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAA ...(((((.......)))))..((((((.(((...((((.((.((((((.....)))))))))))).(((((((....))).))))....)))...))))))(((.........)))... ( -37.20) >consensus ACAGAAUCAGAUUCAGAU______UCGGAUUCAAAGCCGGAGACAGAGUCUGGAACUUUGCUCGGCACGUACGUCACGACGAUAUGAAAAGAGCAAUCGAGUGGCACACAAAAAGCCAAA ........................((((.(((...(((.(((.((((((.....)))))))))))).(((((((....))).))))....)))...)))).((((.........)))).. (-24.10 = -24.10 + -0.00)


| Location | 20,603,038 – 20,603,149 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -18.05 |
| Energy contribution | -20.70 |
| Covariance contribution | 2.65 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20603038 111 - 23771897 AAUGUUUUAGCUUGUACUUUUAUAACUGCAGUCAUGUUAUAUCACCUCUCGACAUGACAUCACCGAUCGAUGGGGUUGGGGUUAUUUCGGGGAGCCGAAAC---AGA------AACAUAA ..(((((..((((.(......((((((...((((((((............))))))))....((((((.....))))))))))))....).))))..))))---)..------....... ( -30.90) >DroSec_CAF1 47935 111 - 1 AAUGUUUUAGCUUGUACUUUUAUAACUGCAGUCAUGUUAUGUCACCUCUCGACAUGACAUCACCGAUCUGCGGGGUUGGGGUUAUUUCGGGGAGCCGAAAC---AGA------AACAUAA ..(((((..((((.(......((((((.((..(((((((((((.......))))))))))..(((.....))))..)).))))))....).))))..))))---)..------....... ( -35.80) >DroAna_CAF1 69914 98 - 1 UAUGUUUCGUCUUGUAGUUUAAUAACUGAAGCCAUGUUAUAUCACCUCCUGACAUGACAUCAUCC-UCAAUAG---------------GGGGAUCCAAAAUAUCAGA------GGGGUAG .((((..((((((.(((((....))))))))..)))..)))).(((((((((.((......((((-((.....---------------)))))).....)).)))).------))))).. ( -22.30) >DroSim_CAF1 42724 111 - 1 AAUGUUUUAGCUUGUACUUUUAUAACUGCAGUCAUGUUAUAUCACCUCUCGACAUGACAUCACCGAUCGGUGGGGUUGGGGUUAUUUCGGGGAGCCGAAAC---AGA------AACAUAA ..(((((..((((.(......((((((.((((((((((............)))))))).(((((....)))))...)).))))))....).))))..))))---)..------....... ( -33.30) >DroEre_CAF1 40371 112 - 1 -ACGUUUCAGCUUGUACUUUUAUGACUGCAGUCAUGUUAUAUCACCUCUCGACAUGACAUCACCGAUCGGCG-GGUUGGGGUUAU---AGGGAGCCGAAAC---AGAAACAGAAACAUAA -..(((((.((((.....(((((((((...((((((((............))))))))....((((((....-))))))))))))---))))))).)))))---................ ( -31.50) >consensus AAUGUUUUAGCUUGUACUUUUAUAACUGCAGUCAUGUUAUAUCACCUCUCGACAUGACAUCACCGAUCGGUGGGGUUGGGGUUAUUUCGGGGAGCCGAAAC___AGA______AACAUAA ...(((((.((((.(......((((((...((((((((............))))))))....((((((.....))))))))))))....).)))).)))))................... (-18.05 = -20.70 + 2.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:25 2006