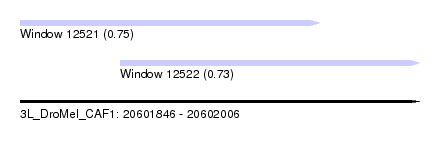
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,601,846 – 20,602,006 |
| Length | 160 |
| Max. P | 0.748517 |

| Location | 20,601,846 – 20,601,966 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.91 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20601846 120 + 23771897 CAAUGGGCAUCUUUAGUGUCCAUCACCCAGCUAAUCCCCAUCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGUGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACG ..((((((((.....))))))))....((((((....(((.....((((((((....(((..(((((..........)))).)..))).)))))))).....)))))))))......... ( -31.50) >DroSec_CAF1 46730 120 + 1 CAAUGGGCAUCUUCAGUGUCCAUCACCCGGCUAAUCCCCAUCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUGUUUGGAGAGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACG ..((((((((.....))))))))....((((((....(((.....((((((((..((..((((.(((..........)))))))..)).)))))))).....)))))))))......... ( -31.20) >DroAna_CAF1 68752 108 + 1 UAAUAGGCAUCUACAGAGUCCAUCACUCU-UAAAUCUCCUUC--UUAAACUUCCUU-------CUCCCAUUUU-UUCGGAGU-CUCAAUGAAGUUUGUUCUUUGGUGGCUGCCAAUUACA ((((.((((.(((((((((.....)))))-............--.((((((((...-------((((......-...)))).-......)))))))).......)))).)))).)))).. ( -24.70) >DroSim_CAF1 41524 120 + 1 CAAUGGGCAUCUUCAGUGUCCAUCACCCGGCUAAUCCCCAUCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGAGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACG ..((((((((.....))))))))....((((((....(((.....((((((((..((..((((.(((..........)))))))..)).)))))))).....)))))))))......... ( -31.20) >DroEre_CAF1 39142 120 + 1 CAAUGGGCAUCUUUAGGGUCCAUCAGCCAGCUAAUCGCCAUCCGUUAAACUGCCCUUUUGUUCCUCCCGUUUUCUUUGGAGUGCUGAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACG ..((((((.((....))))))))..(.((((....(((((.....((((((..(.(((.((..((((.(......).)))).)).))).).)))))).....))))))))).)....... ( -26.70) >consensus CAAUGGGCAUCUUCAGUGUCCAUCACCCAGCUAAUCCCCAUCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGUGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACG ..((((((((.....))))))))....((((((....(((.....((((((((..........((((..........))))........)))))))).....)))))))))......... (-20.63 = -21.91 + 1.28)


| Location | 20,601,886 – 20,602,006 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -27.24 |
| Energy contribution | -26.76 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20601886 120 + 23771897 UCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGUGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACGGCGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGGCAA ..(((((((((((....(((..(((((..........)))).)..))).)))))))((((.((((((((...((((.(((((....))))).))))....))))).))).)))).)))). ( -31.10) >DroSec_CAF1 46770 120 + 1 UCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUGUUUGGAGAGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACGGCGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGGCAA ..(((((((((((..((..((((.(((..........)))))))..)).)))))))((((.((((((((...((((.(((((....))))).))))....))))).))).)))).)))). ( -31.50) >DroAna_CAF1 68791 109 + 1 UC--UUAAACUUCCUU-------CUCCCAUUUU-UUCGGAGU-CUCAAUGAAGUUUGUUCUUUGGUGGCUGCCAAUUACAGUGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGACAA ..--............-------((((......-...)))).-.........((((((((.((((((((...((((.((((......)))).))))....))))).))).)))))))).. ( -24.20) >DroSim_CAF1 41564 120 + 1 UCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGAGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACGGCGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGGCAA ..(((((((((((..((..((((.(((..........)))))))..)).)))))))((((.((((((((...((((.(((((....))))).))))....))))).))).)))).)))). ( -31.50) >DroEre_CAF1 39182 120 + 1 UCCGUUAAACUGCCCUUUUGUUCCUCCCGUUUUCUUUGGAGUGCUGAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACGGCGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGGCAA ..........((((.(((..(((((((.(......).))))....)))..)))..(((((.((((((((...((((.(((((....))))).))))....))))).))).))))))))). ( -30.50) >consensus UCUGUUAAACUUCCUUUUUGUUCCUCCUGUUUUCUUUGGAGUGCUCAAUGAAGUUUGUUCUUUGGUGGCUGUCAAUUACGGCGAGUGUUGUUAUUGUUCUGUCACGUAAUGAGCAGGCAA .......................((((..........))))...........((((((((.((((((((...((((.(((((....))))).))))....))))).))).)))))))).. (-27.24 = -26.76 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:19 2006