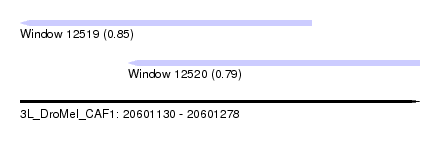
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,601,130 – 20,601,278 |
| Length | 148 |
| Max. P | 0.846541 |

| Location | 20,601,130 – 20,601,238 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
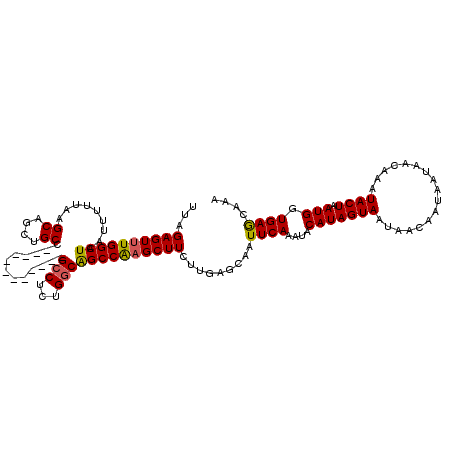
>3L_DroMel_CAF1 20601130 108 - 23771897 UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUCUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAGCAAA ...((((((((((........))).((((((------------(....)))))))))))))).....((...(((....(((((((.................)))).))).)))))... ( -23.63) >DroSec_CAF1 45975 108 - 1 UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUUUGGCAGCCAUGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAGCAAA ((((.((((((((.......(((((((((((------------(....)))))))..)))))....)))..............((....))........))))).))))........... ( -23.10) >DroAna_CAF1 68138 120 - 1 UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCAGCACAGCACACAGCCUCUGACGGCCGAGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAACAAA ...((((((((((............((((.....))))...(((...)))..)))))))))).........((((....(((((((.................)))).))).)))).... ( -22.33) >DroSim_CAF1 40751 108 - 1 UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUUUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAGCAAA ...((((((((((........))).((((((------------(....)))))))))))))).....((...(((....(((((((.................)))).))).)))))... ( -23.63) >DroEre_CAF1 38460 108 - 1 UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUCUGGCGGCCAAGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAACAAA ((((.(((((.......((((((.(((((((------------(((...))))))..)))).))))))...............((....))........))))).))))........... ( -24.20) >consensus UUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC____________GCCUCUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUACAUAGUAAUAACAAUAAUAACAAAUACUAAUGGUGAGCAAA ...((((((((((.........((....)).............(((...))))))))))))).........((((....(((((((.................)))).))).)))).... (-19.41 = -19.17 + -0.24)

| Location | 20,601,170 – 20,601,278 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20601170 108 - 23771897 AGUCCGAGGAGGCCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUCUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUAC .(..((((...(((.(((.((((.....)))))))))).....((((((((((........))).((((((------------(....))))))))))))))))))..)........... ( -34.30) >DroSec_CAF1 46015 108 - 1 AGUCCGAGGAGGCCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUUUGGCAGCCAUGCUUCUUGAGCAAUUCAAAUAC .(..((((((((((.(((.((((.....))))))))))............(((........))).((((((------------(....)))))))....)))))))..)........... ( -32.50) >DroAna_CAF1 68178 117 - 1 ---ACGCGGAGGUCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCAGCACAGCACACAGCCUCUGACGGCCGAGCUUCUUGAGCAAUUCAAAUAC ---.((.((((.((.(((.((((.....)))))))(((.(((((((.((((((........))).((((.....))))...))).))))))).))))).)))).)).............. ( -34.80) >DroSim_CAF1 40791 108 - 1 AGUCCGAGGAGGCCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUUUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUAC .(..((((...(((.(((.((((.....)))))))))).....((((((((((........))).((((((------------(....))))))))))))))))))..)........... ( -34.30) >DroEre_CAF1 38500 108 - 1 AGUCCGAGGAGGCCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC------------GCCUCUGGCGGCCAAGCUUCUUGAGCAAUUCAAAUAC .(..((((...(((.(((.((((.....)))))))))).....((((((((((........)))....(((------------(((...)))))))))))))))))..)........... ( -33.90) >consensus AGUCCGAGGAGGCCUAGAACAAAGUGCAUUUGUCUGGCAGUUAGAGUUUGGCUAAUUUUUAAGCAGCUGCC____________GCCUCUGGCAGCCAAGCUUCUUGAGCAAUUCAAAUAC .(..((((...(((.(((.((((.....)))))))))).....((((((((((.........((....)).............(((...)))))))))))))))))..)........... (-27.40 = -27.64 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:17 2006