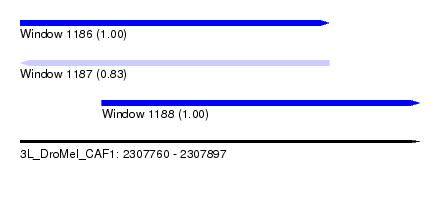
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,307,760 – 2,307,897 |
| Length | 137 |
| Max. P | 0.999935 |

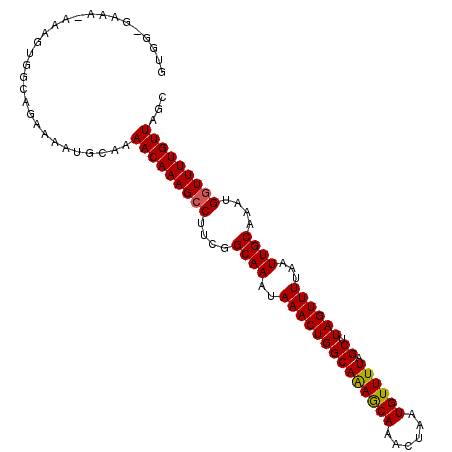
| Location | 2,307,760 – 2,307,866 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.68 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.54 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2307760 106 + 23771897 GUGG-GAAAAAAAGUGUCAGAAAAUGCAAAACAAAGCCUUCGGCAAAUAAACUGGCAAAACAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUCGC (((.-........((((......))))..(((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))))) ( -26.20) >DroSec_CAF1 56928 105 + 1 GUGG-GAAA-AGAGUGGUAGAGAAUGCAAAACAAAGCCUUCGGCAAAUAAACUGGCAGAGCAAGCUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUGGC ....-....-......(((.....)))..(((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))... ( -26.60) >DroSim_CAF1 56875 105 + 1 GUGG-GAAA-AGAGUGGCAGAGAAUGCAAAACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUAGC ....-....-......(((.....)))..(((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))... ( -28.50) >DroEre_CAF1 60168 105 + 1 GCGG-GAAA-AAAGUGGCAAAAAAUUCAAAACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAGUGGUUUUGUUAGU ((.(-....-....).))...........(((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))... ( -26.60) >DroYak_CAF1 59625 105 + 1 GCGGGGAAA-AAAGUGGCAGAAAAUGCUAAACAAAGACUUCGGCAAAUAAACUGGCAAAACAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAGUGCUUUUGUUAG- .(((((...-....(((((.....)))))........)))))(((((...(((.((((...(((((((.((....)))))))))...)))).)))...)))))...- ( -24.16) >consensus GUGG_GAAA_AAAGUGGCAGAAAAUGCAAAACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUAGC .............................(((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))... (-24.74 = -24.54 + -0.20)

| Location | 2,307,760 – 2,307,866 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.68 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2307760 106 - 23771897 GCGAACAAAACCAUUUGCAAUUAAAACUAAGCUAAAACAUUAGUUUGUUUUGCCAGUUUAUUUGCCGAAGGCUUUGUUUUGCAUUUUCUGACACUUUUUUUC-CCAC (((((((((.((.((.((((...(((((..((.((((((......)))))))).)))))..)))).)).)).)))).)))))....................-.... ( -23.10) >DroSec_CAF1 56928 105 - 1 GCCAACAAAACCAUUUGCAAUUAAAACUAAGCUAAAACAUUAGCUUGCUCUGCCAGUUUAUUUGCCGAAGGCUUUGUUUUGCAUUCUCUACCACUCU-UUUC-CCAC ((.((((((.((.((.((((...((((((((((((....)))))))((...)).)))))..)))).)).)).))))))..))...............-....-.... ( -20.10) >DroSim_CAF1 56875 105 - 1 GCUAACAAAACCAUUUGCAAUUAAAACUAAGCUAAAACAUUAGUUUGCUUUGCCAGUUUAUUUGCCGAAGGCUUUGUUUUGCAUUCUCUGCCACUCU-UUUC-CCAC ((.((((((.((.((.((((...(((((((((..((((....))))))))....)))))..)))).)).)).))))))..))...............-....-.... ( -19.30) >DroEre_CAF1 60168 105 - 1 ACUAACAAAACCACUUGCAAUUAAAACUAAGCUAAAACAUUAGUUUGCUUUGCCAGUUUAUUUGCCGAAGGCUUUGUUUUGAAUUUUUUGCCACUUU-UUUC-CCGC .....((((((.(((.((((...(((((((.........)))))))...)))).)))......(((...)))...))))))................-....-.... ( -17.90) >DroYak_CAF1 59625 105 - 1 -CUAACAAAAGCACUUGCAAUUAAAACUAAGCUAAAACAUUAGUUUGUUUUGCCAGUUUAUUUGCCGAAGUCUUUGUUUAGCAUUUUCUGCCACUUU-UUUCCCCGC -..((((((..(....((((...(((((..((.((((((......)))))))).)))))..))))....)..))))))..(((.....)))......-......... ( -17.70) >consensus GCUAACAAAACCAUUUGCAAUUAAAACUAAGCUAAAACAUUAGUUUGCUUUGCCAGUUUAUUUGCCGAAGGCUUUGUUUUGCAUUUUCUGCCACUUU_UUUC_CCAC ...((((((.((.((.((((...((((((((((((....)))))))((...)).)))))..)))).)).)).))))))............................. (-14.90 = -14.58 + -0.32)

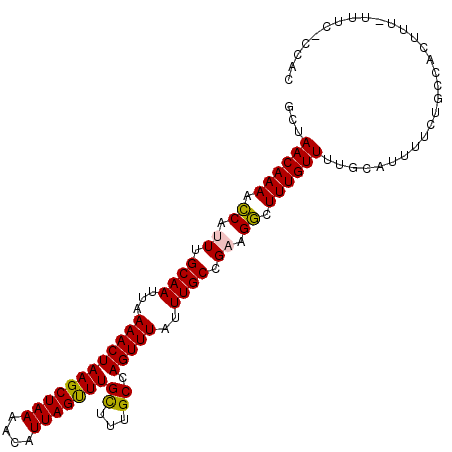
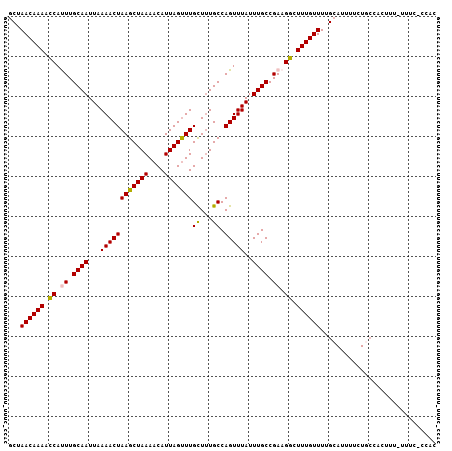
| Location | 2,307,788 – 2,307,897 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2307788 109 + 23771897 AACAAAGCCUUCGGCAAAUAAACUGGCAAAACAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUCGCGUUAGGCAAUCCAAGCGUCUGAA-----UUAUAUGC---- (((((((((....((((..((((((((((((((......)))))).)).))))))...))))....))))))))).(((((.((....)).))))).....-----........---- ( -29.90) >DroSec_CAF1 56955 109 + 1 AACAAAGCCUUCGGCAAAUAAACUGGCAGAGCAAGCUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUGGCGUUAGGCAAUCCAAGCGUCUGAA-----UUAUAUGC---- (((((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))))((((((.((....)).))))))....-----........---- ( -33.40) >DroSim_CAF1 56902 109 + 1 AACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUAGCGUUAGGCAAUCCAAGCGUCUGAA-----UUAUAUGC---- (((((((((....((((..((((((((((((((......)))))).)).))))))...))))....))))))))).(((((.((....)).))))).....-----........---- ( -30.50) >DroEre_CAF1 60195 115 + 1 AACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAGUGGUUUUGUUAGUGUUGGGCAAUCCAAAGAUCUGAA---AUUUACUCGUAUGC ..(((((((....((((..((((((((((((((......)))))).)).))))))...))))....)))))))((((..((((.....))))....)))).---.............. ( -28.20) >DroYak_CAF1 59653 109 + 1 AACAAAGACUUCGGCAAAUAAACUGGCAAAACAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAGUGCUUUUGUUAG------GGAAACCAAAGAUAUAUGCUCAAUUAUUCGUU--- ((((((..(....((((..((((((((((((((......)))))).)).))))))...))))....)..)))))).(------(....)).........................--- ( -23.70) >consensus AACAAAGCCUUCGGCAAAUAAACUGGCAAAGCAAACUAAUGUUUUAGCUUAGUUUUAAUUGCAAAUGGUUUUGUUAGCGUUAGGCAAUCCAAGCGUCUGAA_____UUAUAUGC____ (((((((((....((((..((((((((((((((......)))))).)).))))))...))))....))))))))).(((((.((....)).)))))...................... (-26.20 = -26.84 + 0.64)

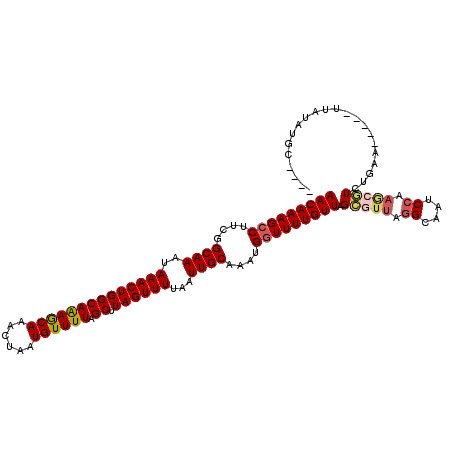
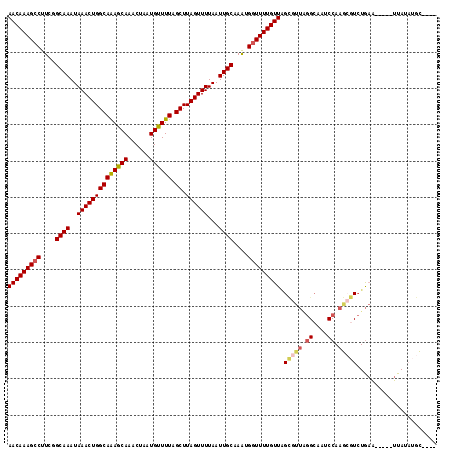
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:49 2006