
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
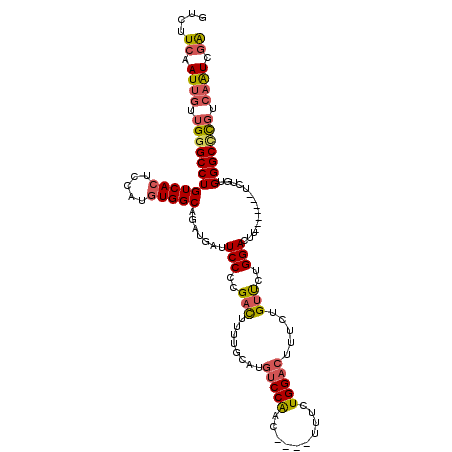
| Location | 20,599,750 – 20,599,859 |
| Length | 109 |
| Max. P | 0.718828 |

| Location | 20,599,750 – 20,599,859 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -24.64 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
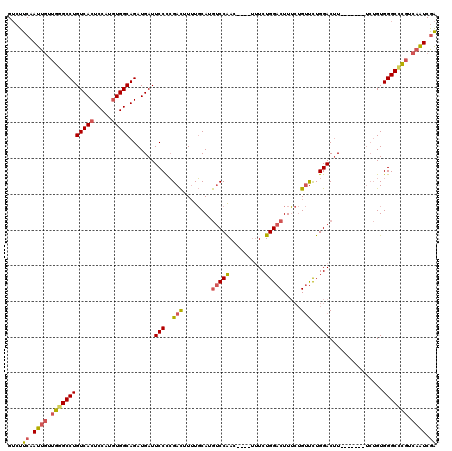
>3L_DroMel_CAF1 20599750 109 + 23771897 GUCUUCAAUUGUUGGGCCUGUCACUCCAUGUGGCAGAUGAUUCCCCGACUUUUGCAUGUCCAAC----UUUCUGGACUUUCUGUUCUGGACUU-------UCUGUGGGCCCGUCAAUCGA ....((.((((.(((((((..(((.....)))(((((....(((..(((........(((((..----....))))).....)))..)))...-------)))))))))))).)))).)) ( -34.32) >DroSec_CAF1 44461 109 + 1 GUCUUCAAUUGUUGGGCCUGUCACUCCAUGUGGCAGAUGAUUCCCCGACUUUUGCAUGUCCAAC----UUUCUGGACUUUCUGUUCUGGACUU-------UCUGUGGGCCCGUCAAUCGA ....((.((((.(((((((..(((.....)))(((((....(((..(((........(((((..----....))))).....)))..)))...-------)))))))))))).)))).)) ( -34.32) >DroAna_CAF1 66759 118 + 1 GGCUUCAAUUAUUGGGCCUGUCACUCCAGCUGGCAGAUGAUUCCUGGAUAUAUCCAUAACCGAUCCAUGGUCUGGACUUG--GACUUGGACUUGGACUGGACAGUGGGCUUUUAUGUCAG (((((........)))))...((((((((...............((((....))))...((((((((.(((((......)--)))))))).)))).)))))..)))(((......))).. ( -32.10) >DroSim_CAF1 37899 109 + 1 GUCUUCAAUUGUUGGGCCUGUCACUCCAUGUGGCAGAUGAUUCCCCGACUUUUGCAUGUCCAAC----UUUCUGGACUUUCUGUUCUGGACUU-------UCUGUGGGCCCGUCAAUCGA ....((.((((.(((((((..(((.....)))(((((....(((..(((........(((((..----....))))).....)))..)))...-------)))))))))))).)))).)) ( -34.32) >DroEre_CAF1 37047 108 + 1 GUCUUCAAUUGUUGCGCCUGUCACUCCAUGUGGCAGAUGAUUCCCCGACUUUUGCAUGUCCAAC----UUUCUGGACUUUCUGUUCUGGACAU-------U-UGUGGGCCCGUCAAUCGA ....((.((((.((.((((..(((.....)))(((((((..(((..(((........(((((..----....))))).....)))..))))))-------)-))))))).)).)))).)) ( -27.92) >consensus GUCUUCAAUUGUUGGGCCUGUCACUCCAUGUGGCAGAUGAUUCCCCGACUUUUGCAUGUCCAAC____UUUCUGGACUUUCUGUUCUGGACUU_______UCUGUGGGCCCGUCAAUCGA ....((.((((.((((((((((((.....))))).......(((..(((........(((((..........))))).....)))..)))...............))))))).)))).)) (-24.64 = -25.36 + 0.72)

| Location | 20,599,750 – 20,599,859 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
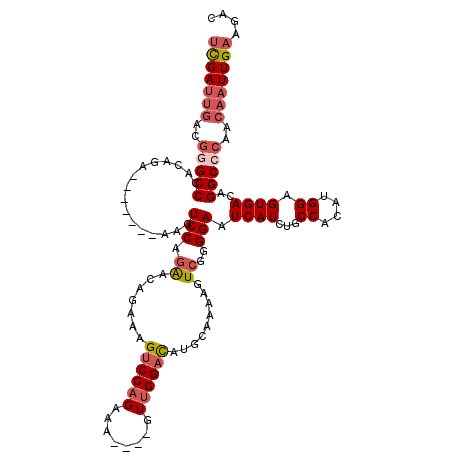
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -21.95 |
| Energy contribution | -23.47 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
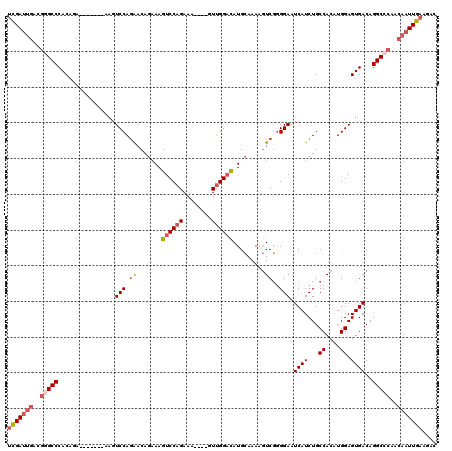
>3L_DroMel_CAF1 20599750 109 - 23771897 UCGAUUGACGGGCCCACAGA-------AAGUCCAGAACAGAAAGUCCAGAAA----GUUGGACAUGCAAAAGUCGGGGAAUCAUCUGCCACAUGGAGUGACAGGCCCAACAAUUGAAGAC (((((((..(((((..((((-------...(((.((.......((((((...----.)))))).........))..)))....)))).(((.....)))...)))))..))))))).... ( -34.09) >DroSec_CAF1 44461 109 - 1 UCGAUUGACGGGCCCACAGA-------AAGUCCAGAACAGAAAGUCCAGAAA----GUUGGACAUGCAAAAGUCGGGGAAUCAUCUGCCACAUGGAGUGACAGGCCCAACAAUUGAAGAC (((((((..(((((..((((-------...(((.((.......((((((...----.)))))).........))..)))....)))).(((.....)))...)))))..))))))).... ( -34.09) >DroAna_CAF1 66759 118 - 1 CUGACAUAAAAGCCCACUGUCCAGUCCAAGUCCAAGUC--CAAGUCCAGACCAUGGAUCGGUUAUGGAUAUAUCCAGGAAUCAUCUGCCAGCUGGAGUGACAGGCCCAAUAAUUGAAGCC ...........((((((..((((((....(((((...(--(..(((((.....))))).))...))))).....((((.....))))...)))))))))...)))............... ( -26.40) >DroSim_CAF1 37899 109 - 1 UCGAUUGACGGGCCCACAGA-------AAGUCCAGAACAGAAAGUCCAGAAA----GUUGGACAUGCAAAAGUCGGGGAAUCAUCUGCCACAUGGAGUGACAGGCCCAACAAUUGAAGAC (((((((..(((((..((((-------...(((.((.......((((((...----.)))))).........))..)))....)))).(((.....)))...)))))..))))))).... ( -34.09) >DroEre_CAF1 37047 108 - 1 UCGAUUGACGGGCCCACA-A-------AUGUCCAGAACAGAAAGUCCAGAAA----GUUGGACAUGCAAAAGUCGGGGAAUCAUCUGCCACAUGGAGUGACAGGCGCAACAAUUGAAGAC (((((((..(.((((((.-.-------...((((.........((((((...----.))))))........((.(.(((....))).).)).)))))))...))).)..))))))).... ( -28.10) >consensus UCGAUUGACGGGCCCACAGA_______AAGUCCAGAACAGAAAGUCCAGAAA____GUUGGACAUGCAAAAGUCGGGGAAUCAUCUGCCACAUGGAGUGACAGGCCCAACAAUUGAAGAC (((((((..(((((................(((.((.......((((((........)))))).........))..))).((((...((....)).))))..)))))..))))))).... (-21.95 = -23.47 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:15 2006