
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,599,082 – 20,599,195 |
| Length | 113 |
| Max. P | 0.723562 |

| Location | 20,599,082 – 20,599,195 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20599082 113 + 23771897 CGAUGGCAGUGGGCUUCACUUGGCUUUACUCCGGGUUCUCGG-GCUCGCCUCUGUAGCGAUGGGGCAUGCCCCAAAAGCCG--GGCUUCUGGCCCACUCCCAG----UCUUUCCCCGAUC .(((((.((((((((...(((((((((..(((((....))))-).((((.......))))(((((....))))))))))))--)).....)))))))).))).----))........... ( -48.00) >DroSec_CAF1 43811 113 + 1 CGAUGGCAGUGGGCUUCACUUGGCUAUACUCCGGGUUCUCGG-GCUCGCCUCUCUAGCGAUGGGGCAUGCCCCAAAAGCCG--GGCUUCUGGCCCACUCCCAG----UCUUUCCCCGAUC .(((((.((((((((...(((((((....(((((....))))-).((((.......))))(((((....)))))..)))))--)).....)))))))).))).----))........... ( -46.20) >DroAna_CAF1 66091 100 + 1 CGACGUCAGUGGGCUUCACUUGGUG--------------------UCUCGUCUGUGCCGAAGGGGCAUGCCCCCAAAGCUAUUGCCUUUUGGCCCACUCCCAGGUCUUCUUUCCCCGGGC .......((((((((.....(((..--------------------(.......)..)))((((((((.((.......))...))))))))))))))))(((.((.........)).))). ( -31.00) >DroSim_CAF1 37249 113 + 1 CGAUGGCAGUGGGCUUCACUUGGCUUUACUCCGGGUUCUCGG-GCUCGCCUCUCUAGCGAUGGGGCAUGCCCCAAAAGCCG--GGCUUCUGGCCCACUCCCAG----UCUUUCCCCGAUC .(((((.((((((((...(((((((((..(((((....))))-).((((.......))))(((((....))))))))))))--)).....)))))))).))).----))........... ( -48.00) >DroEre_CAF1 36395 114 + 1 CGAUGGCAGUGGGCUUCACUUGGCUGUACUCCGAGUUCUCGGGGCUCACCUCUCUAGCGAUGGGGCCUGCCCCAAAAGACA--GGCUUCUGGCCCACUCCCAG----UCUUUCCCCGAUC .(((((.((((((((((.((.((.((..((((((....))))))..)))).....)).)).((((((((.(......).))--)))))).)))))))).))).----))........... ( -43.40) >consensus CGAUGGCAGUGGGCUUCACUUGGCUUUACUCCGGGUUCUCGG_GCUCGCCUCUCUAGCGAUGGGGCAUGCCCCAAAAGCCG__GGCUUCUGGCCCACUCCCAG____UCUUUCCCCGAUC ...(((.((((((((.....(((((.....((((....))))..................(((((....)))))..))))).........)))))))).))).................. (-30.38 = -30.90 + 0.52)

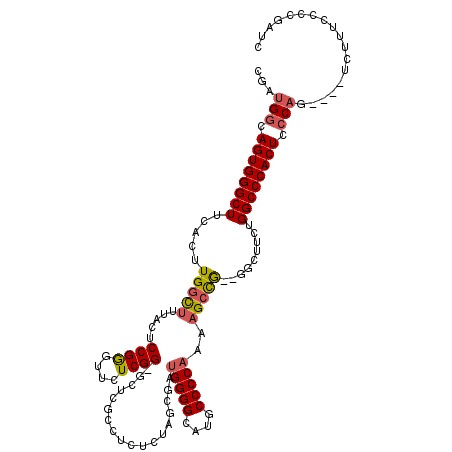
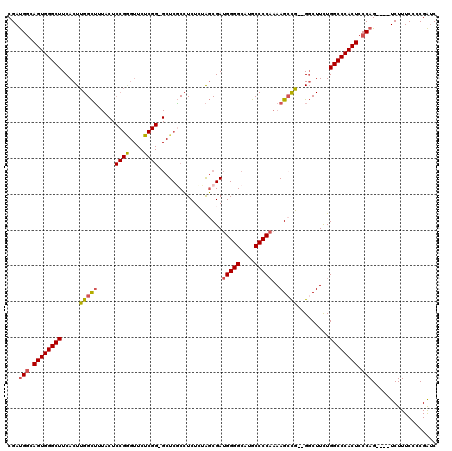
| Location | 20,599,082 – 20,599,195 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -45.34 |
| Consensus MFE | -35.45 |
| Energy contribution | -36.34 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20599082 113 - 23771897 GAUCGGGGAAAGA----CUGGGAGUGGGCCAGAAGCC--CGGCUUUUGGGGCAUGCCCCAUCGCUACAGAGGCGAGC-CCGAGAACCCGGAGUAAAGCCAAGUGAAGCCCACUGCCAUCG ...........((----.(((.(((((((.....((.--.(((((((((((....)))))(((((.....)))))((-(((......))).))))))))..))...))))))).))))). ( -47.10) >DroSec_CAF1 43811 113 - 1 GAUCGGGGAAAGA----CUGGGAGUGGGCCAGAAGCC--CGGCUUUUGGGGCAUGCCCCAUCGCUAGAGAGGCGAGC-CCGAGAACCCGGAGUAUAGCCAAGUGAAGCCCACUGCCAUCG ...........((----.(((.(((((((.....(((--(((.((((.((((.((((...((....))..)))).))-)).)))).)))).)).....(....)..))))))).))))). ( -46.70) >DroAna_CAF1 66091 100 - 1 GCCCGGGGAAAGAAGACCUGGGAGUGGGCCAAAAGGCAAUAGCUUUGGGGGCAUGCCCCUUCGGCACAGACGAGA--------------------CACCAAGUGAAGCCCACUGACGUCG .((((((.........))))))(((((((............(((..(((((....)))))..)))..........--------------------(((...)))..)))))))....... ( -38.60) >DroSim_CAF1 37249 113 - 1 GAUCGGGGAAAGA----CUGGGAGUGGGCCAGAAGCC--CGGCUUUUGGGGCAUGCCCCAUCGCUAGAGAGGCGAGC-CCGAGAACCCGGAGUAAAGCCAAGUGAAGCCCACUGCCAUCG ...........((----.(((.(((((((.....((.--.(((((((((((....)))))(((((.....)))))((-(((......))).))))))))..))...))))))).))))). ( -47.10) >DroEre_CAF1 36395 114 - 1 GAUCGGGGAAAGA----CUGGGAGUGGGCCAGAAGCC--UGUCUUUUGGGGCAGGCCCCAUCGCUAGAGAGGUGAGCCCCGAGAACUCGGAGUACAGCCAAGUGAAGCCCACUGCCAUCG ...........((----.(((.(((((((.....(((--(((((....))))))))....(((((.....(((..((.((((....)))).))...))).))))).))))))).))))). ( -47.20) >consensus GAUCGGGGAAAGA____CUGGGAGUGGGCCAGAAGCC__CGGCUUUUGGGGCAUGCCCCAUCGCUAGAGAGGCGAGC_CCGAGAACCCGGAGUAAAGCCAAGUGAAGCCCACUGCCAUCG ..................(((.(((((((..(((((.....)))))(((((....)))))(((((.....(((..((.(((......))).))...))).))))).))))))).)))... (-35.45 = -36.34 + 0.89)

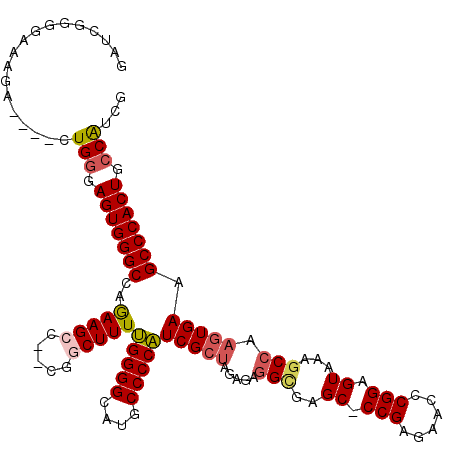
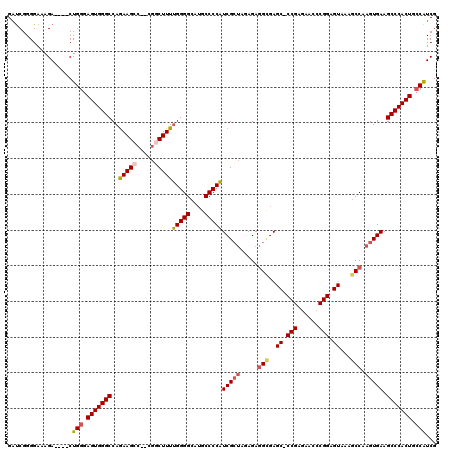
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:12 2006